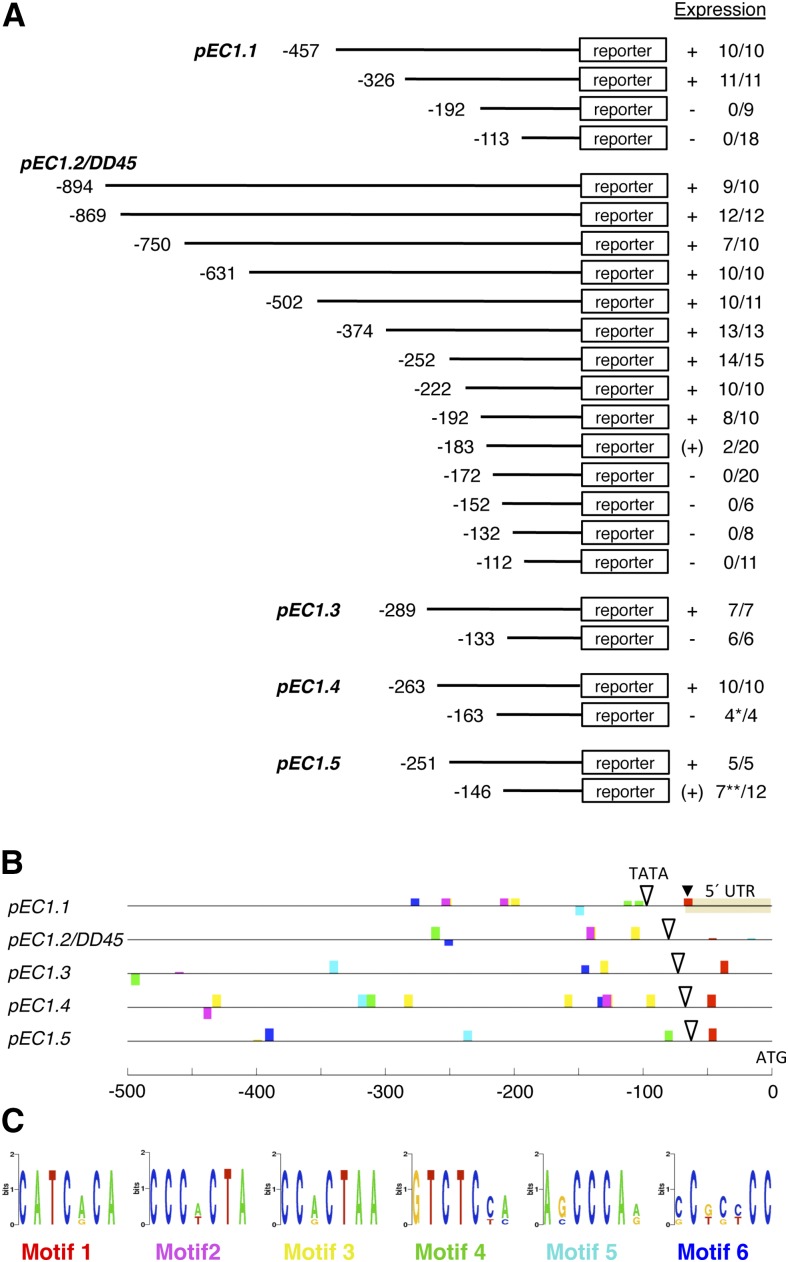
Figure 2.
EC1 promoter deletion studies and mapping of putative cis-regulatory motifs. A, Scheme summarizing the results from EC1 promoter deletion studies. A series of 5′ deletion constructs was tested for reporter activity in transgenic plants. Expression describes the observed reporter activity as present (+), weakly present [(+)], or absent (−) in the egg cell. Numbers indicate individual transgenic lines for a given deletion construct showing reporter activity compared with the total number of lines transgenic for this construct. *, One out of four lines showed misexpression of the reporter in sporophytic cells; **, five of seven lines showed only very weak reporter activity, and two of these five lines showed misexpression in sporophytic cells. B, Conserved sequence motifs (colored boxes) mapped in the −500-bp upstream regions of the five EC1 genes by Cistome (https://bar.utoronto.ca/cistome/cgi-bin/BAR_Cistome.cgi) using the prediction program MEME. White triangles mark the positions of TATA box motifs identified by AthMap (http://www.athamap.de/index.php). The transcription start site of EC1.1 is labeled with a black triangle. UTR, Untranslated region. C, Sequence logos of mapped sequence motifs shown in B. Motifs 2 and 3 show high sequence similarity.