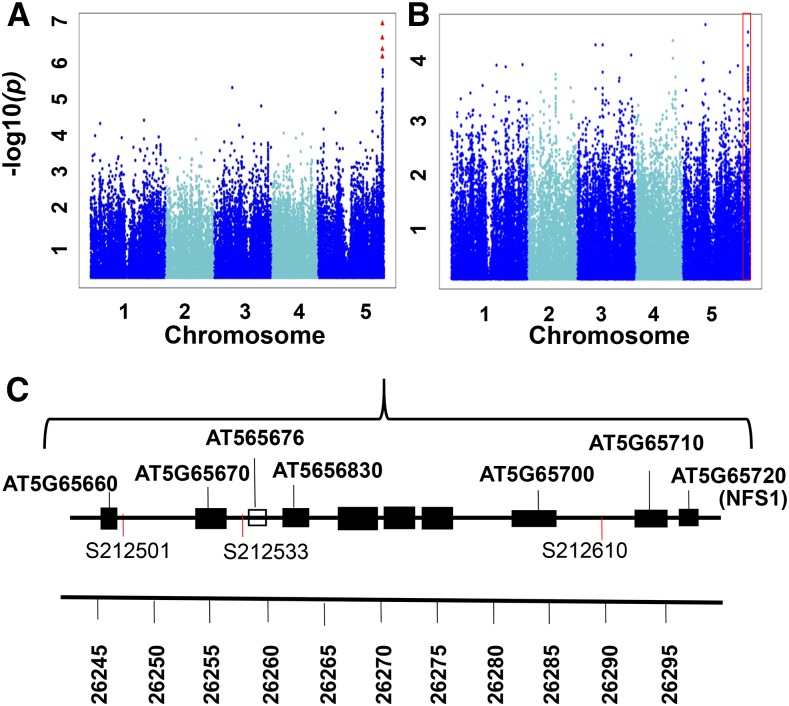
Figure 2.
GWAS summary of S/LIVKHSQR. A and B, Scatterplots of the association results from a unified mixed model analysis of S/LIVKHSQR (A) and S/total (B) traits across the five Arabidopsis chromosomes. The negative log10-transformed P values from the GWAS analysis are plotted against the genomic physical positions. P values for SNPs that are statistically significant for a trait at 5% FDR are in red. The red box represents the corresponding chromosomal region of this significant signal in the S/total Manhattan plot. C, Graphical representation of the genes within the vicinity of SNP212501, SNP212533, and SNP212610. Genes are represented by black boxes, and a pseudogene is represented by the white box. At5G65720 encodes a Cys desulfurase (NFS1) that is the leading candidate gene for S/LIVKHSQR.