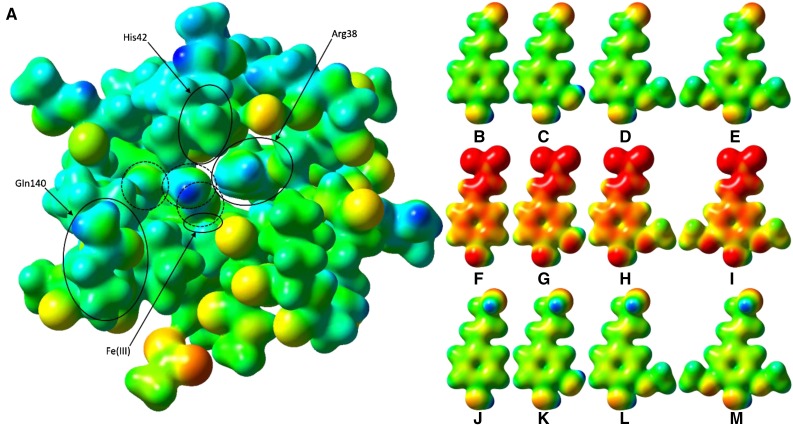
Figure 10.
Electrostatic potential surfaces of PviPRX9 and phenylpropanoids. A, Heme (hexcoordinate, high spin), Arg-31 (as methylguanidinium), Ser-35 (as ethanol), Arg-38 (as n-propylguanidinium), Phe-41 (as toluene), His-42 (as 5-methyl-1H-imidazole; mostly obscured by Leu-66), Ala-65 (as ACE) to Arg-73 (as NMA), Gly-170 (as formamide), Asp-135 (as ACE) to Phe-141 (as NMA), Ala-172 (as ACE) to Phe-177 (as NMA), His-167 (as 5-methyl-1H-imidazole), Asp-326 (as acetate ion), and six water molecules; Cys-174 was mutated to Gly to reduce computational cost. ACE and NMA are acetyl and N-methyl capping groups on the N and C termini, respectively, of each stretch of multiple residues. The three water molecules circled with dashed lines were, from right to left, the water involved in the first proton transfer, a noncatalytic water that occupies the nascent binding site of the hydroxycinnamyl substrate phenol functional group, and the water released upon H2O2 binding to the heme. The latter water molecule was regenerated by heterolytic cleavage of H2O2 and involved in catalytic proton transfer. B, p-Coumaryl aldehyde. C, Caffeyl aldehyde. D, Coniferyl aldehyde. E, Sinapyl aldehyde. F, p-Coumarate. G, Caffeate. H, Ferulate. I, Sinapate. J, p-Coumaryl alcohol. K, Caffeyl alcohol. L, Coniferoyl alcohol. M, Sinapyl alcohol. The active site and ligand electrostatic potentials, mapped on their corresponding self-consistent field densities, are shown on a potential scale of −2.7 × 10−1 hartrees (red) to +2.97 × 101 hartrees (blue) for the active site and −1 × 10−1 (red) to 3 × 101 (blue) for the ligands. This figure was generated using GaussView 3.09.