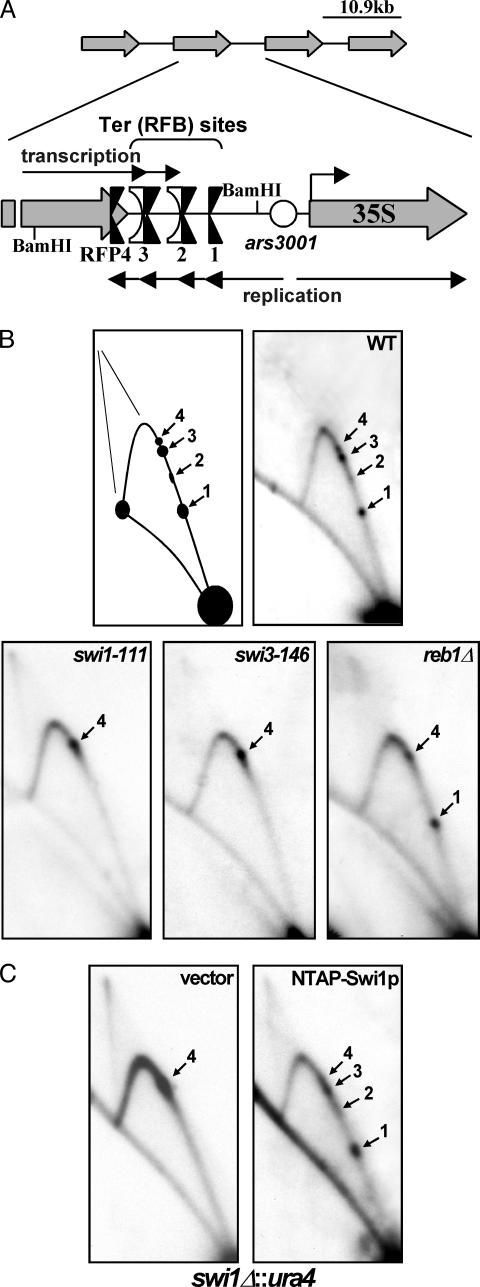
Fig. 1.
2D gel autoradiograms of replication intermediates of genomic rDNA showing the topology of fork movement in the region. (A) Diagram depicting the array of tandemly repeated rDNA units located on both arms of chromosome III. A closer view of the intergenic region shows the presence of the four rDNA fork barriers Ter (RFB) 1–3 and RFP4 near the 3′ end of the 35S transcription unit. Note the presence of RFP4 upstream of Reb1p binding sites Ter2 and Ter3, which represent two of three in vivo transcription terminators. (B) Autoradiograms of 2D gel analyses of the replication intermediates of the BamHI fragment depicted in A. Ter1–Ter3 and RFP4 localize to the ascending Y-arc of this fragment in WT cells (black arrows). Ter1–Ter3 are absent in swi1-111 and swi3-146 cells, whereas only Ter2 and Ter3 are absent in reb1Δ::kanMX cells. Note that forks accumulate at RFP4 in the swi1 and swi3 (and reb1Δ) mutants. (C) TAP-tagged Swi1p fully complements swi1Δ cells at Ter1–Ter3. Two-dimensional gel analysis of a swi1Δ::ura4 strain (SPGK301C) expressing only the TAP epitope (vector) or N-terminally tagged Swi1p (NTAP-Swi1p).