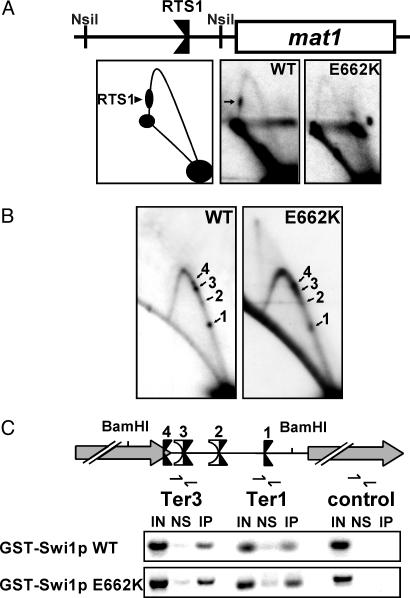
Fig. 4.
2D gel analysis of fork movement at the mating-switch locus and ChIP analyses of the rDNA Ter sites. The E662K mutant form of Swi1p, which is completely defective in fork arrest at RTS1, is fully functional at the rDNA fork barriers and localizes to this region as effectively as WT Swi1p. (A) 2D gel analysis of the centromere-proximal NsiI fragment of mat1 containing RTS1, confirming results in ref. 7 that Swi1p E662K fails to arrest forks at this site. RTS1 (arrow) can be seen on the descending Y-arc in WT cells but not in cells harboring the swi1 E662K mutation (E662K). A diagram of the analyzed region is shown. (B) Ter1–Ter3 are functional in swi1 E662K mutant cells. 2D gel analysis of the intergenic BamHI fragment of WT and swi1 E662K (E662K) cells. (C) ChIP. WT GST-Swi1p localizes to the rDNA Ter sites as shown by enrichment of this region as compared with a region within the rDNA transcription unit located ≈5 kb from the fork barriers. Consistent with 2D replication analysis (see B), GST-Swi1p E662K localizes to the Ter sites at least as efficiently as the WT protein. Note that this assay cannot clearly resolve the closely associated Ter sites. A diagram of the analyzed region is shown.