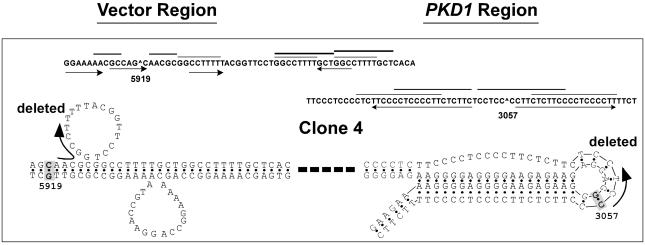
Fig. 3.
Representative DNA structures for clone 4. The nucleotides in boldface and shading correspond to homologous sequences. The arrows indicate the sequences that were deleted, and a thick dashed line denotes intervening DNA. Repetitive motifs are identified as in the legend to Fig. 7. The breakpoint in the PKD1 tract (G·C 3057) occurred within a 7-bp stretch that was flanked by two perfect 19-bp mirror repeats (TTCCCCTCCCCTTCTCTTC). The mirror symmetry and the R·Y composition are consistent with the formation of an intramolecular triplex with a DNA loop of 7 bp. The location of the breakpoint within the loop coincides with nucleotides known to have unusual torsion angles (26) and to be susceptible to chemical modification and nuclease cleavage in vitro. In the vector, the breakpoint (G·C 5919) was located 6 bp upstream of three direct repeats, GGCCTTTT followed after 9 bp by two tandem GGCCTTTTGCT repeats. In the looped structure resulting from slippage with the first repeat, the breakpoint is located in a region of perturbed helical DNA.