FIGURE 2.
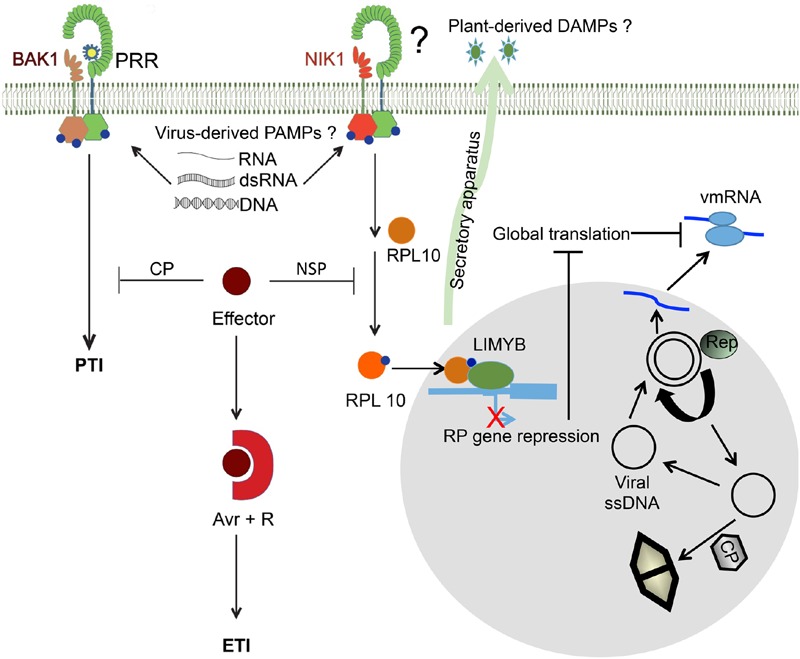
Similarities between viral PTI and NIK1-mediated antiviral signaling. Replication and expression of viral genomes lead to the accumulation of non-self DNA or RNA motifs (virus-derived PAMPs), which may be recognized by PRRs that in turn heteromultimerize with co-receptors (BAK1 or SERK1) to trigger viral PTI. Alternatively, PTI may be activated by endogenous DAMPs, which are induced by virus infection and delivered to the apoplast via the secretory apparatus. In addition to PTI, in the case of DNA viruses (begomoviruses), plant cells may also elicit the translational control branch of the NIK1-mediated antiviral signaling as an innate defense. The mechanism of NIK1 transmembrane receptor activation is unknown. Structural organization and biochemical properties of NIK1 may suggest an activation mechanism dependent on recognition of viral PAMPs or endogenous DAMPs by PRR partners, similarly to a typical viral PTI. In this case, one may consider virus derived-dsDNA as possible PAMPs. The viral single-stranded DNA form begomoviruses replicates via double-stranded DNA intermediates that are transcribed in the nucleus of plant-infected cells. NSP binds to the nascent viral DNA and facilitates its movement to the cytoplasm and acts in concert with the classical MP to transport the viral DNA to the adjacent, uninfected cells. Activation of NIK1 in incompatible interactions promotes phosphorylation and subsequent translocation of RPL10 to the nucleus, where it interacts with LIMYB to fully repress the expression of RP genes, leading to global translation suppression, which also impairs viral mRNA (vmRNA) translation. In begomovirus-host compatible interactions, NSP binds to NIK1 and suppresses its activity. In any case, RNA or DNA viruses, a successful infection implicates in accumulation of virus effectors (for example, CP from PPV and NSP from begomoviruses) to suppress PTI, leading to disease. In resistant genotypes, however, the resistance genes specifically recognize, directly or indirectly, the viral effectors, called avirulence (Avr) factors, activating ETI and conferring resistance. Adapted from Machado et al. (2015).
