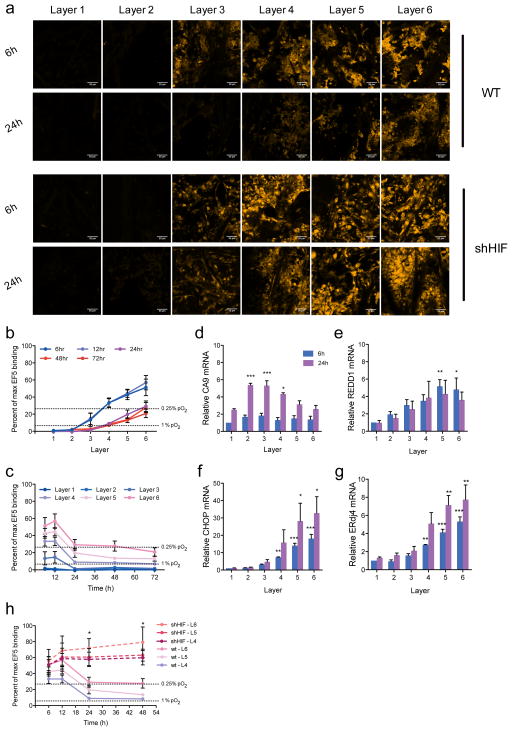
Figure 3. Oxygen gradients within the TRACER over time.
a- EF5 (yellow) staining in layers 1–6 of the TRACER after 6h, and 24h in WT and shHIF cells. All scale bars 100 microns. Gradients in EF5 staining (and hence oxygen levels) are clearly visible at 6 and 24h. b – Percentage of maximum EF5 binding (inversely correlated with oxygen levels) in WT cells quantified from nuclear EF5 fluorescence in each layer between 6 and 72h in culture. c - Percentage of maximum EF5 binding in WT cells as a function of time. Error bars in b-c are SEM (n=3). Dashed lines indicate conversions to hypoxia levels based on previously reported correlations30. d–g -Quantitative reverse transcriptase–PCR analysis of HIF target genes CA9 and REDD1 and the UPR response genes CHOP and ErDj4 relative to housekeeping gene RPL13a in each layer (L1 (outer) to L6 (inner)) after 6h (blue bar) and 24h (magenta bars). Data normalized to 6h, layer 1. Error bars are standard deviation (n=3 TRACERs). P-values obtained with one-way ANOVA, Dunnett post-test. *P<0.05, **P<0.01,***P<0.001. Single layer controls provided in Supplementary Figure 14. h - Percentage of maximum EF5 binding as a function of time for TRACERs containing WT (solid lines) or shHIF (dashed lines) cells. Error bars are SEM (n=3).