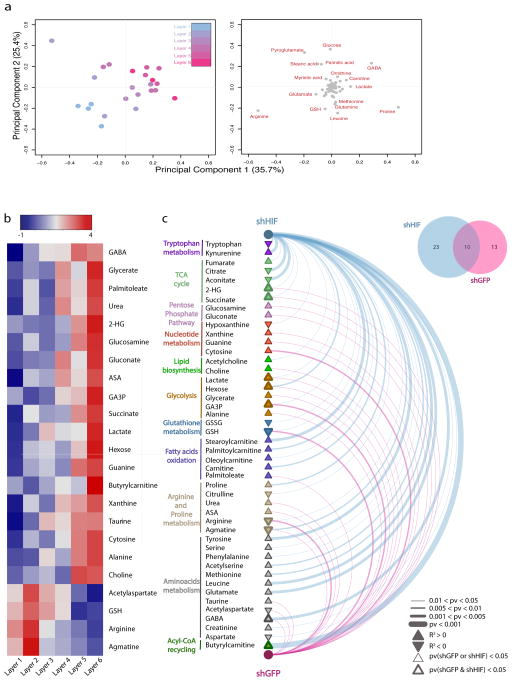
Figure 4. Layer-specific metabolomic analysis and correlation with hypoxia in the TRACER.
a - Principal component analysis (PCA) of the metabolomic signature of cells in the indicated TRACER layers. Score plot (left) and loading plot (right) of principal components 1 and 2 are indicated. Samples in the score plot are color-coded according to the layer of origin. Metabolites that covariate with samples clustering are highlighted in red in the loading plot. b -Heatmap representation of metabolite intensities per layer for metabolites that show a significant correlation (Pearson pv < 0.05) with hypoxia levels. Hypoxia levels reported in Figure 3 were linearly transformed by applying a probit transformation (see methods) and Pearson correlation analysis was performed to test for non-zero correlations between the transformed hypoxia levels and metabolite intensities in each layer. c - Arc plot of metabolites showing significant correlation with hypoxia across the TRACER containing shGFP cells (violet) and shHIF cells (blue). Line thickness is indicative of p-value significance. Metabolite nodes are represented as filled triangles, where pathway are colour coded and the border thickness indicates whether each metabolite was found significant in shGFP or shHIF alone (thin border) or both in shGFP and shHIF (thick border). The Venn diagram in the top right corner indicates the overlap proportion of significant metabolites in shGFP and shHIF. Measurements are generated from n = 4 TRACERs.