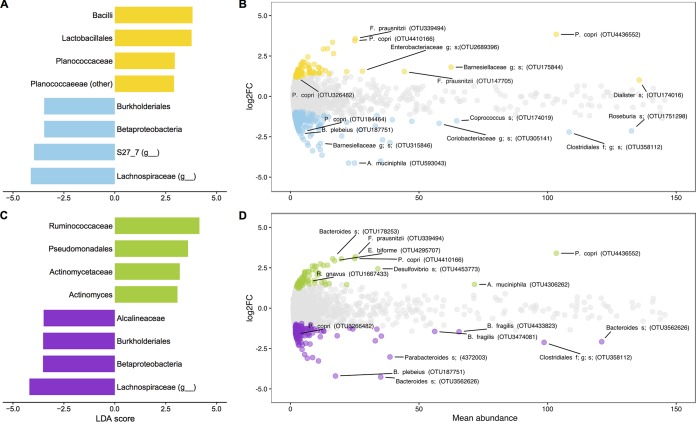
FIG 4 .
Differentially abundant OTUs and higher taxonomic units across geography and diet. (A and C) Linear discriminant analyses (LDA) using LEfSe were applied to identify biomarkers at higher taxonomic levels (down to the genus level). (B and D) Differentially abundant OTUs were identified using DESeq2 (see Materials and Methods). (A and B) Samples were compared across geographic regions (for Montreal, n = 26 [in yellow]; for Nunavut, n = 19 [in blue]) for LEfSe biomarkers (A) and differentially abundant OTUs (B) identified by DESeq2. (C and D) Samples were compared by diet (for the Western diet, n = 29 [in green]; for the Inuit diet, n = 19 [in purple]) for LEFSe biomarkers (C) and differentially abundant OTUs (D) identified by DESeq2. All associations had P values of <0.05 after correction for multiple tests. Only the data from the top four LEfSe biomarkers (LDA score of >2.5) for each category are presented here. For full LEfSe and DESeq2 results, see Tables S1D to G and Fig. S6A to C. The differentially abundant OTUs named as indicated in panels B and D focus on those discussed in the main text.