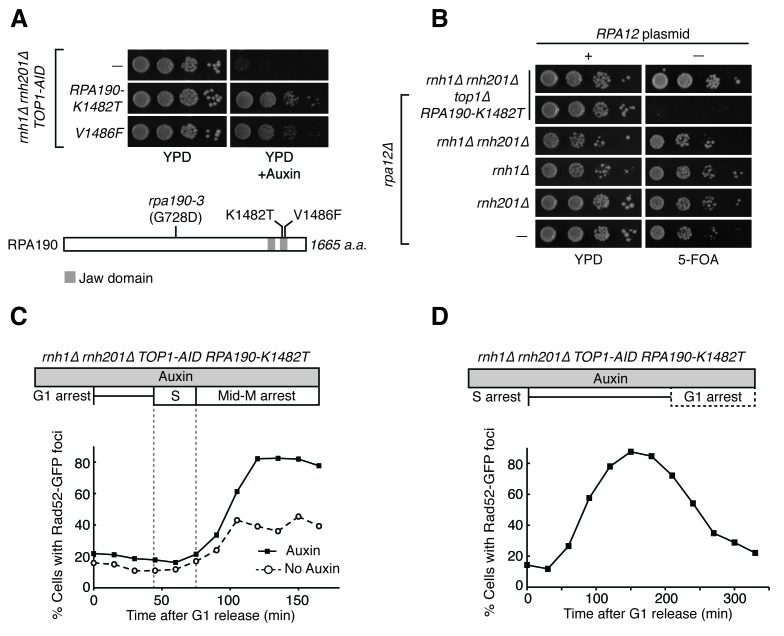
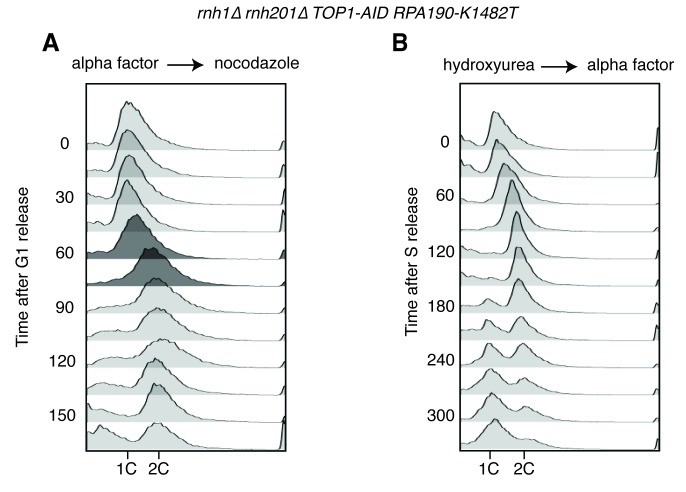
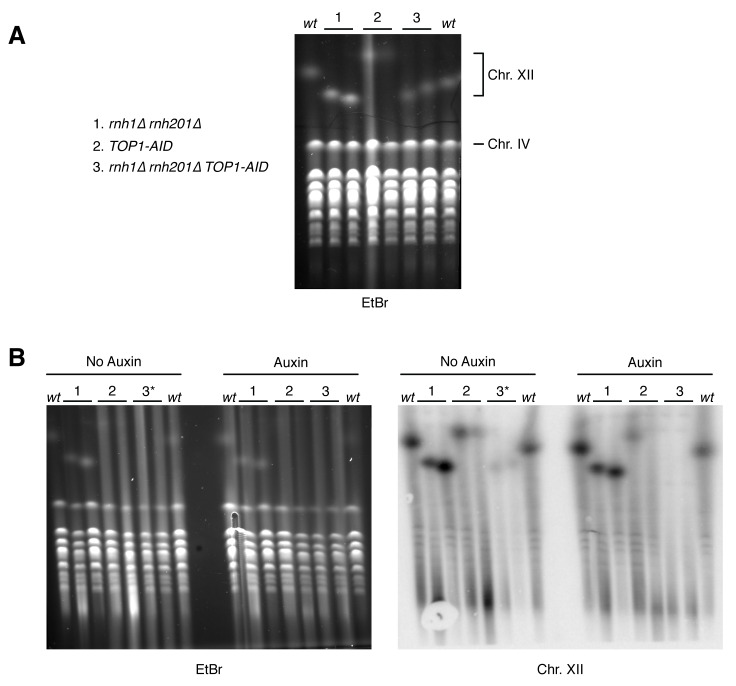
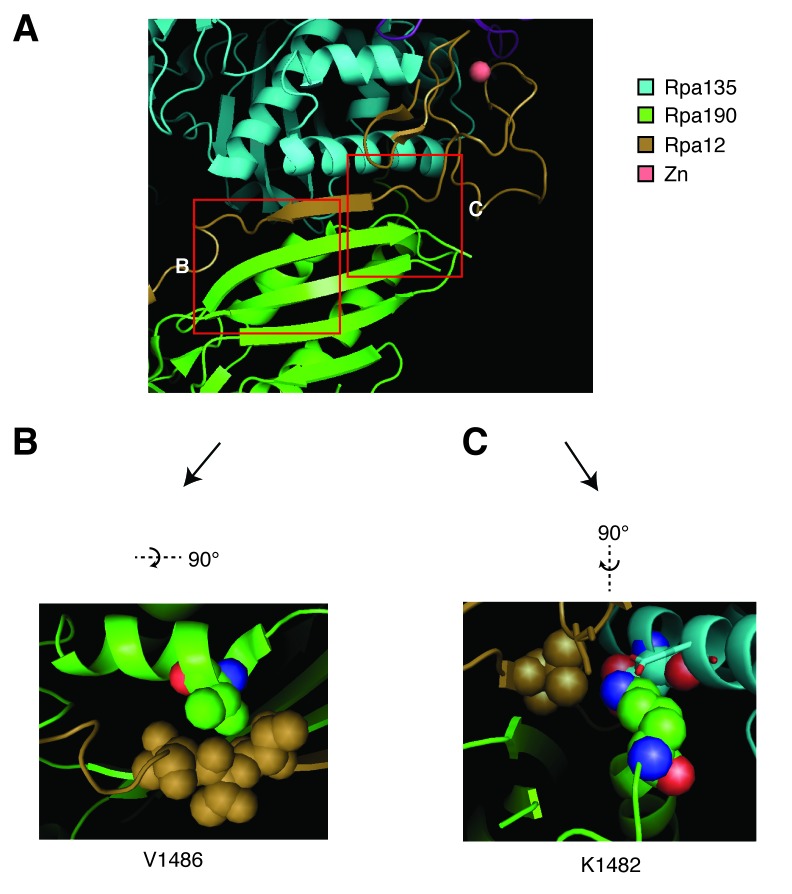
Figure 7. RPA190 mutants allow for repair of R-loop induced damage.
(A) Top: Rpa190-K1482T and -V1486F suppress auxin sensitivity of rnh1∆ rnh201∆ TOP1-AID cells. 10-fold serial dilutions of saturated cultures were plated onto YPD or YPD with auxin. Bottom: Schematic of Rpa190 showing the location of mutations and the jaw domain, as previously published (Engel et al., 2013; Fernández-Tornero et al., 2013). (B) Rpa12 is required in rnh1∆ rnh201∆ TOP1-AID RPA190-K1482T. Cells carrying a plasmid expressing RPA12 and URA3 were plated onto media lacking uracil (-URA, selects for plasmid) or media containing 5-floroorotic acid (5-FOA, selects for plasmid loss). 10-fold serial dilutions are shown. (C) Rpa190-K1482T does not change accumulation of Rad52-GFP foci. Experiment in Figure 4A was repeated on rnh1∆ rnh201∆ TOP1-AID RPA190-K1482T cells. (D) Rpa190-K1482T allows for repair of Rad52-GFP foci. Experiment in Figure 2B was repeated on rnh1∆ rnh201∆ TOP1-AID RPA190-K1482T cells in the presence of auxin.