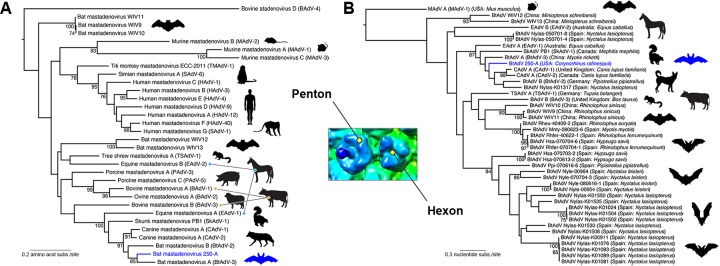
FIG 6.
Maximum-likelihood phylogenetic analysis of BtAdV 250-A with other mastadenoviruses. For both the penton and the hexon trees, bootstrap values (>70%) are shown for key nodes. The structural location of the penton and hexon proteins in the BtAdV capsid are shown in the central inset. BtAdV 250-A is highlighted by a blue branch and bat in each tree for reference. (A) Phylogenetic tree of the amino acid sequences of the penton protein for member species of the genus Mastadenovirus (i.e., from a diversity of mammalian taxa). The tree is midpoint rooted for clarity only, and horizontal branch lengths are scaled according to the number of amino acid substitutions per site. GenBank accession numbers for the sequences used in the penton tree were as follows: BAdV-1 (YP_094036), BAdV-2 (NP_597922), BAdV-3 (AEW91335), BAdV-4 (NP_077393), BtAdV-2 (YP_004782104), BtAdV-3 (YP_005271186), BtAdV 250-A (KX871230), BtAdV WIV9 (KT698853), BtAdV WIV10 (KT698854), BtAdV WIV11 (KT698855), BtAdV WIV12 (KT698856), BtAdV WIV13 (KT698852), CAdV-1 (NP_044193), CAdV-2 (AP_000617), EAdV-1 (AEP16413), EAdV-2 (YP_009162351), HAdV-1 (AP_000507), HAdV-3 (YP_002213774), HAdV-4 (AAW33302), HAdV-9 (YP_001974428), HAdV-12 (NP_040920), HAdV-40 (NP_040857), MAdV-1 (NP_015541), MAdV-2 (YP_004123742), MAdV-3 (YP_002822211), PAdV-3 (YP_009205), PAdV-5 (NP_108662), SAdV-1 (YP_213970), SAdV-6 (AFG19591), SkAdV-1 (YP_009162592), TMAdV (YP_007518317), and TSAdV-1 (YP_068064). (B) Evolutionary relationships between BtAdV 250-A and BtAdVs from Europe and Asia were assessed using nucleotide sequences of the hexon gene. Closely related viruses (CAdV-1, CAdV-2, SkAdV-1, EAdV-1, EAdV-2, TSAdV-1, and BAdV-3) as determined from the penton phylogeny in panel A, were also included in the analysis. The tree is rooted using the more divergent sequence of Murine mastadenovirus A (MAdV A), and horizontal branch lengths are scaled according to the number of nucleotide substitutions per site. GenBank accession numbers for the sequences used in the hexon tree were as follows: BAdV-3 (AF030154), BtAdV-2 (JN252129), BtAdV-3 (NC_016895), BtAdV 250-A (KX871230), BtAdV WIV9 (KT698853), BtAdV WIV10 (KT698854), BtAdV WIV11 (KT698855), BtAdV WIV12 (KT698856), BtAdV WIV13 (KT698852), CAdV-1 (AC_000003), CAdV-2 (AC_000020), EAdV-1 (JN418926), EAdV-2 (KT160425), MAdV-1 (M81889), SkAdV-1 (KP238322), TSAdV-1 (AF258784), and BtAdV population set HM856327 to HM856353 (courtesy of I. Casas, S. Vazquez, J. Juste, A. Falcon, C. Aznar, C. Ibanez, F. Pozo, G. Ruiz, J. M. Berciano, I. Garin, J. Aihartza, P. Perez-Brena, and J. E. Echevarria, National Centre of Microbiology, Madrid, Spain).