FIG 1.
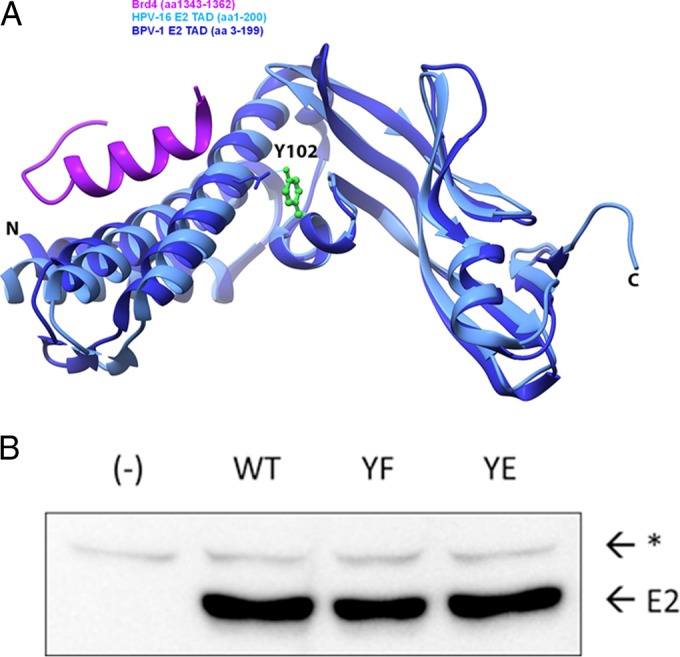
Location of Y102 in the TAD and mutagenesis. (A) A ribbon diagram of the BPV-1 E2 TAD crystal structure reported under PDB accession number 2JEU (60) (dark blue) overlapping the HPV-16 E2 TAD/Brd4 cocrystal structure reported under PDB accession number 2NNU (38) (light blue and purple, respectively) was created by using the UCSF Chimera package. The Y102 side chain, depicted in a green ball-and-stick model, is hydrogen bonded to E74, depicted in a wire frame model. N, amino terminus; C, carboxyl terminus. (B) Site-directed mutagenesis converts Y102 to phenylalanine (F) or glutamate (E) in pCG-E2. One microgram of WT and mutant constructs (YF and YE), or mRFP-GFP as negative control (−), was expressed in HEK293TT cells. The immunoblot was probed for BPV-1 E2 with mouse monoclonal antibody B201. E2, full-length BPV-1 E2; *, nonspecific band.
