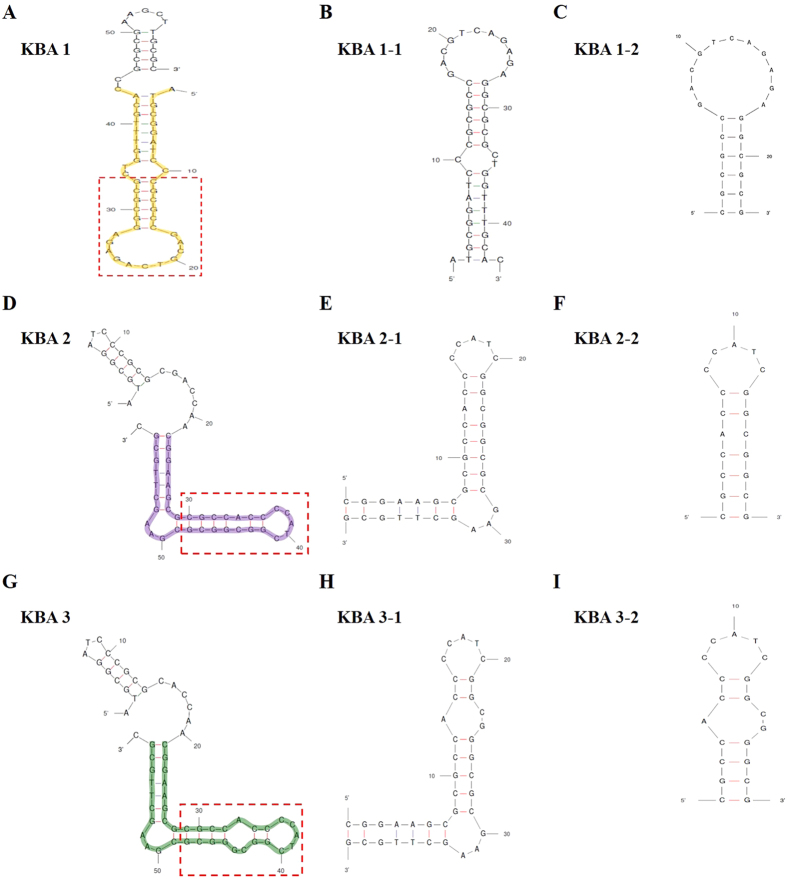
Figure 2. Schematic representation of the secondary structures of selected kanamycin binding aptamers (KBAs) and its truncation form generated by the M-fold program.
(A–C) Derivatives of KBA 1, (D–F) derivatives of KBA 2, and (G–I) derivatives of KBA 3. 5′ or 3′ primer binding sites and several unpaired terminal nucleotides were eliminated, and the highlighted region indicates the first truncation form (B,E and H). Additional truncations of stem-loop motifs are shown in the red dot box (C,F and I).