Abstract
Background
Oral colostrum priming (OCP) after birth in preterm infants is associated with improved weight gain and modification of the oral immuno-microbial environment. We hypothesized OCP would modify salivary immune peptides and the oral microbiota in preterm infants.
Methods
We conducted a prospective, randomized clinical trial to determine the effects of OCP on salivary immune peptide representation in preterm infants (<32 weeks completed gestation at birth). Saliva samples were collected prior to and after OCP. Salivary immune peptide representation was determined via mass spectroscopy. Oral microbiota representation was determined via sequencing of 16S rRNA gene.
Results
Neonates that received OCP (n = 48) had a 16-day reduction in the median length of hospitalization as compared to infants that did not receive OCP (n = 51). No differences in salivary immune peptide sequence representation prior to OCP between groups were found. Longitudinal changes in peptides were detected (lysozyme C, immunoglobulin A, lactoferrin) but were limited to a single peptide difference (alpha defensin 1) between primed and unprimed infants after OCP. We found no difference in microbial diversity between treatment groups at any time point, but diversity decreased significantly over time in both groups. OCP treatment marginally modified oral taxa with a decline in abundance of Streptococci in the OCP group at 30 days of life.
Conclusions
OCP had neither an effect on the salivary peptides we examined nor on overall oral bacterial diversity and composition. Infants that received OCP had a reduced length of hospitalization and warrants further investigation.
Keywords: preterm, neonate, colostrum, oral priming, saliva, microbiome, proteome
Introduction
Human milk ingestion is associated with a decreased risk of developing infection and necrotizing enterocolitis in preterm infants1, 2. Breast milk contains multiple functional immunologic elements that contribute to improved neonatal host immune defense3, 4. Recent work has shown that mixing of milk and the neonate’s saliva appears to represent a unique biochemical synergism that may boost early innate immunity5. Breast milk feeding is associated with an altered oral and nasopharyngeal microbiota later in infancy6, 7. However, oral feeding is not practical in most preterm infants due to developmental immaturity8. Recently, the practice of early (as soon as produced by the mother) oral administration of mother’s own colostrum was proposed as a protective strategy9. Like breast feeding in term infants, recent studies have shown OCP may influence the microbial colonization of the oral cavity in very low birth weight infants (<1500g, VLBWs)10, decrease clinical sepsis, inhibit secretion of pro-inflammatory cytokines, increase levels of circulating immune-protective factors in extremely preterm infants (<28 weeks)11, and increase weight at 36 weeks corrected age12.
While OCP appears a safe, cost-effective, and easily implementable practice that may improve overall weight gain, it remains unclear if oral colostrum priming changes the oral immuno-microbial milieu or overall health in very preterm newborns12. Also remaining unclear are whether these effects are passive and thus must be continued, or if priming results in temporary or sustained changes. We hypothesized OCP would modify salivary immune peptides and the oral microbiota in preterm infants.
Methods
Human subjects and sample processing
We performed a prospective, randomized trial of OCP in preterm infants and examined the impact over time on salivary immunopeptides and also examined the oral microbiota in a subset of these patients. The study began February 2013 and finished July 2014. The Institutional Review Board at Vanderbilt University approved the study prior to its initiation. Inclusion criteria were birth at <32 weeks completed gestation. Exclusion criteria were not meeting inclusion criteria, decline to participate, enrollment in competing studies, or Spanish-speaking only. Following informed consent from a parent, preterm infants (<32 weeks completed gestation) were randomized by a numeric list generated by Dr. Slaughter a priori to receive oral priming with mother’s colostrum or no oral priming. Participating staff members were not blinded. The rate of breast-feeding intention among mothers of infants in the neonatal intensive care unit at Monroe Carell Jr. Children’s Hospital at Vanderbilt exceeded 95% during the period of study. Maternal and neonatal demographics as well as birth hospitalization outcomes were collected and analyzed.
Oral priming with mother’s colostrum
Orogastric gavage feeding was not restricted in patients of either group during the period of colostrum priming. Freshly pumped maternal colostrum (mother’s first milk production after birth) was aliquoted and kept refrigerated at 4°C until administered. Neonates randomized to OCP received 100 µL of colostrum in each cheek via a 1ml syringe every 6 hours for 5 days beginning in the first 48 hours of life similar to our previous era-based study12. Aliquoted colostrum to be used over the first 24 hours (4 syringes) was refrigerated at 4°C and was never frozen. The policy at Vanderbilt was to keep milk refrigerated (before or after thawing) for a maximum of 24 hours so remaining aliquots were frozen at −20°C until the day of use.
Saliva collection
Neonatal saliva was obtained at 3 time points: day of life (DOL) 1–2, DOL 8–9 and DOL 30. These time points were selected a priori to 1) minimize the potential impact of residual amniotic fluid/lung liquid in the mouth, 2) allow for collection of samples 24–48 hours after the completion of oral priming, and 3) obtain data from a fixed post-priming time point for microbiota determination. To collect saliva, a wicking sterile sponge at the end of an applicator shaft (Beaver Visitec) was inserted directly into to the neonate’s mouth using sterile technique for a minimum of 60 seconds, removed and immediately sealed into a sterile 15ml conical tube (Thermo Fisher). If adequate saliva for analysis could not be collected, 100 µL of sterile 0.9% saline was used to irrigate the mouth and a second wicking sponge was used for collection. Saliva or oral washing was isolated from the sponge via centrifugation, sterilely aliquoted for proteomic or microbiota studies, and stored at −80°C until processed in batches.
Salivary peptide measurement
Representative peptides for each protein were selected based on their appearance across samples in the discovery phase of analysis in which series of unscheduled runs in saliva samples retention times and the most useful transitions to monitor (Supplemental Figure 1). Heavy-labeled peptide internal standards were synthesized by JPT (SpikeTides TQL peptides, JPT, Berlin, Germany) that contained isotopically-labeled terminal arginine and lysine residues (13C and 15N) and a trypsin-cleavable C-terminal tag. These isotopically-labeled peptides were spiked into samples at approximately endogenous levels prior to digestion. Skyline software (University of Washington, MacCoss lab) was used to set up scheduled, targeted MRM methods and three to five MRM transitions were monitored per peptide. Additional methods are in Supplemental methods.
Oral bacterial DNA isolation
Saliva samples for oral microbiota determinations were obtained using sterile technique. DNA was extracted using a modified Qiagen protocol as previously described13. Briefly, 50 µl of saliva was pretreated with 20 mg/ml lysozyme in TRIS-HCl and EDTA buffer (Qiagen, DNeasy Blood & Tissue Kit, catalog no. 69504) for lysis of Gram-positive bacteria. The remainder of the bacterial DNA extraction protocol proceeded per the manufacturer’s instructions. All DNA samples were evaluated for quality using spectrophotometry and agarose gel electrophoresis. To control for environmental contamination of study samples, bacterial DNA was extracted from template-free negative controls and sequenced in parallel with patient samples as described below.
16S rRNA amplicon sequencing of bacterial DNA
Total DNA (12.5 ng) was amplified using a combination (4:1) of universal and Bifidobacterium-specific primers targeting the V1–V2 region of the bacterial 16S rRNA gene14, 15. Master mixes contained 12.5 ng of total DNA, 0.2 µM of each primer and 2× KAPA HiFi HotStart ReadyMix (KAPA Biosystems, Wilmington, MA). Primer sequences contained overhang adapters appended to the 5’ end of each primer for compatibility with the Illumina sequencing platform (Supplemental methods). Additional methods are in Supplemental methods.
Sequencing data analysis
Multiplexed paired-end fastq files were produced from the sequencing results of the Illumina MiSeq using the Illumina software configureBclToFastq. The paired-end fastqs were joined into a single multiplexed, single-end fastq using the software tool fastq-join. Demultiplexing and quality filtering was performed on the joined results. Quality analysis reports were produced using the FastQC software. Bioinformatics analysis of bacterial 16S rRNA amplicon sequencing data was conducted using the Quantitative Insights Into Microbial Ecology (QIIME) software16. OTU picking was performed on the quality filtered results using pick_de_novo_otus.py. Chimeric sequences were detected and removed using ChimeraSlayer17. Alpha diversity and beta diversity analysis were performed on the data set using the QIIME routines: alpha_rarefaction.py and beta_diversity_through_plots.py18, 19, respectively. Summary reports of taxonomic assignment by sample and all categories were produced using QIIME summarize_taxa_through_plots.py and summarize_otu_by_cat.py.
Statistical approaches and analytical methods
Prior to conducting the study, power calculations were performed assuming 50 subjects in each group. For comparing the priming to no priming groups, we had 90% power to detect our primary outcome, which was a 0.46 standard deviation differences in peptide means between groups. Secondary endpoints of interest were length of stay, total days intubated, age at feeding initiation, days to 100ml/kg/day of enteral feeds, as well as rates of necrotizing enterocolitis and sepsis. For analyses of changes over time, the effect size we were able to detect depended on the correlation between the DOL 1–2 and DOL 8–9 measurements. If the correlation was high, r=0.8, then would have 90% power to detect a 0.13 standard deviation change in mean over time. If the correlation was lower, we would have 90% power to detect a 0.33 (r=0.5) or 0.52 (r=0.2) standard deviation change in mean over time. All tests have a two-tailed type I error probability of 0.05.
Continuous variables were summarized using the median, 25th, and 75th percentiles, and categorical variables were summarized using percentages. We used the non-parametric Wilcoxon rank sum to test if continuous variables differed by randomization group, and we used Pearson Chi-squared test for categorical variables. All subjects were analyzed according to their randomization assignment (intent to treat). The Wilcoxon signed rank test for paired day was used to compare peptide levels on DOL 1–2 to levels on DOL 8–9. Spearman's rank correlation was used to test for associations among peptides. Analysis of similarity tests (ANOSIM) were applied to specific categories in the metadata mapping file to separate samples into groups, and then test whether there were significant differences between those groups. R and P values were calculated using the phylogeny-based unweighted UniFrac distance metric. For identification of bacterial taxa differentially represented in specific categories we compared OTU frequencies in sample groups using a nonparametric ANOVA test (Kruskal Wallis). P-values were corrected for multiple comparisons by the Benjamini-Hochberg FDR procedure. P-values of less than 0.05 were considered significant.
Results
Patient Characteristics and Outcomes
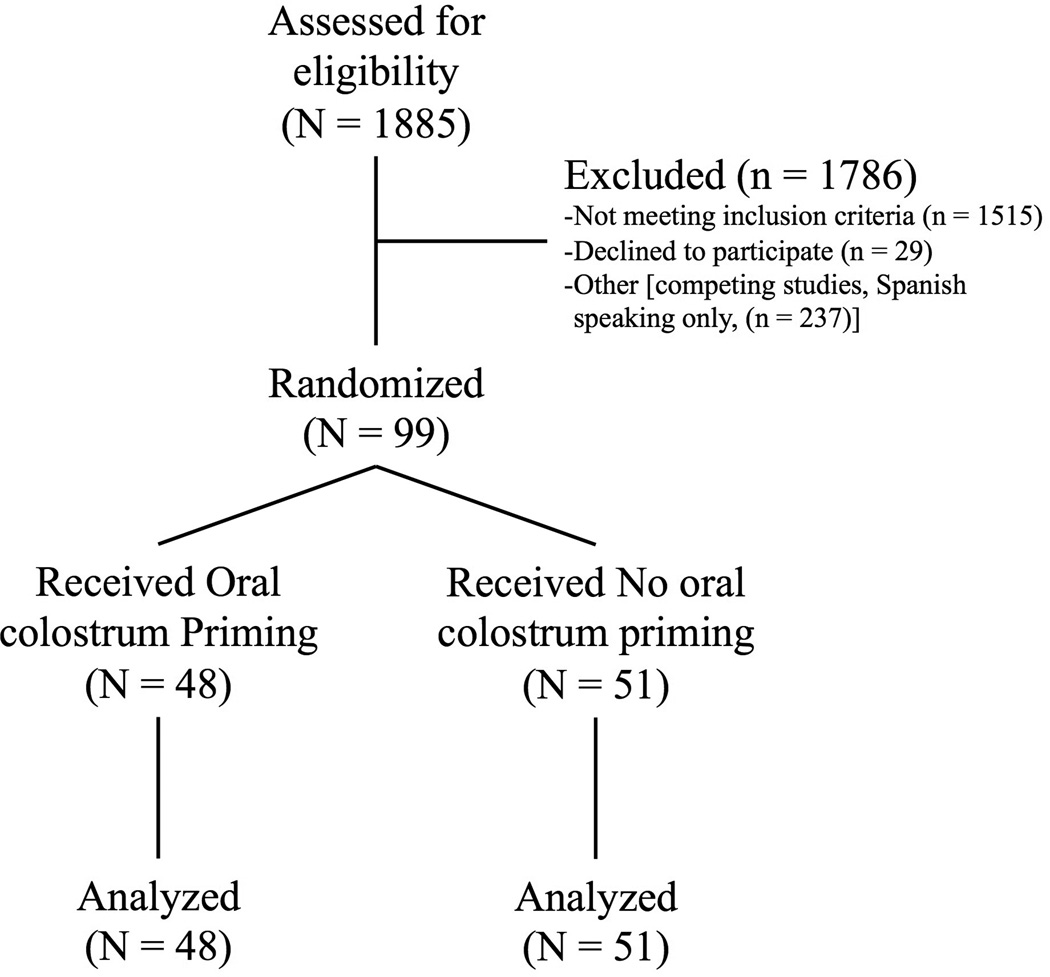
The details of the enrollment, allocation, and analysis are shown in Figure 1. The two groups of patients were well-matched in maternal and neonatal characteristics (Table 1). Mothers of neonates randomized to OCP had a greater incidence of prenatal care (100% OCP versus 92% no OCP, P=0.045 by Pearson) and diabetes (17% OCP versus 4% no OCP, P=0.035 by Pearson). Necrotizing enterocolitis (stage≥2) occurred in 1 patient in the no OCP group and in 2 patients in the OCP group (P=0.64 by Pearson). A sixteen-day reduction in median length of hospitalization was noted for the infants that received oral colostrum priming (40 days) as compared to infants that did not (56 days, p = 0.04). Of note, this reduction in length of hospitalization was not accompanied by a reduced time to 100 ml/kg/day of enteral feedings among infants that were primed compared to infants who were not primed (p=0.40). The number of days from the last dose of caffeine recorded to the day of discharge [median duration 20 days (No OCP) vs 22 (OCP), p = 0.41] did not support the hypothesis that the OCP group had a reduced number of events that might have shortened the length of hospitalization. We also measured the duration from attainment of full PO feeds to discharge [6 days (No OCP) vs 5 days (OCP), p = 0.55], and the time to full PO feeds [47 days (No OCP) vs 39 days (OCP), p = 0.25]. There was only one death in the cohort (in the OCP group). No adverse events were noted among patients in either group during the course of the study.
Figure 1. CONSORT diagram.
Table 1.
Maternal and neonatal characteristics
| No OCP | OCP | P-value | ||
|---|---|---|---|---|
| Maternal Characteristics | N = 51 | N = 48 | ||
| Age, median (quartiles) | 26 (21, 30) | 25 (23, 29) | 0.571 | |
| Race | ||||
| African American | 25% | 40% | 0.372 | |
| White | 69% | 58% | ||
| More than one | 4% | 2% | ||
| Unknown, not reported | 2% | 0% | ||
| Ethnicity | 0.682 | |||
| Hispanic or Latino | 14% | 17% | ||
| Parity, median (quartiles) | 2 (1, 2) | 2 (1, 2) | 0.421 | |
| Gestation (multiple) | 20% | 19% | 0.912 | |
| Prenatal care (n=98) | 92% | 100% | 0.0452 | |
| Mode of delivery | 0.532 | |||
| SVD | 25% | 31% | ||
| Caesarian section | 75% | 69% | ||
| Anesthesia | 0.492 | |||
| Epidural | 75% | 83% | ||
| General | 16% | 8% | ||
| Hypertension (n=98) | 29% | 38% | 0.352 | |
| Diabetes | 4% | 17% | 0.0352 | |
| Tobacco use(n=97) | 16% | 21% | 0.52 | |
| Premature rupture of membranes* | 25% | 25% | 0.952 | |
| Amniotic fluid description (n=98) | ||||
| Clear | 84% | 91% | 0.492 | |
| Bloody | 14% | 6% | ||
| Meconium | 2% | 2% | ||
| Antenatal steroids | 88% | 90% | 0.832 | |
| Neonatal characteristics | ||||
| Gestational age (weeks), median (quartiles) | 29 (28, 30) | 30 (27, 31) | 0.252 | |
| Birth weight (grams), median (quartiles) | 1170 (905–1340) | 1272 (988, 1602) | 0.121 | |
| Male | 41% | 50% | 0.382 | |
| 5 minute Apgar score, median (quartiles) | 7 (6, 8) | 7 (6, 8) | 0.741 | |
| Surfactant | 59% | 71% | 0.212 | |
| Total days intubated | 1 (0, 5) | 1 (0, 3) | 0.971 | |
| Early onset bacteremia | 2% | 0% | 0.332 | |
| Late onset bacteremia | 6% | 2% | 0.342 | |
| Antimicrobial exposure, days, median (quartiles) | 7 (3, 9) | 3 (2, 8) | 0.111 | |
| Age at feeding initiation (days), median (quartiles) | 2 (2, 3) | 2 (2, 3) | 0.671 | |
| Initial feeding type | 0.972 | |||
| Expressed breast milk | 69% | 71% | ||
| Donor breast milk | 29% | 27% | ||
| Formula | 2% | 2% | ||
| Days to 100ml/kg/day enteral feeds, median (quartiles) |
11 (9, 19) | 11 (8, 15) | 0.391 | |
| Type of feeds at discharge | ||||
| Unfortified breast milk | 4% | 6% | 0.12 | |
| Fortified breast milk | 41% | 60% | ||
| Formula only | 55% | 33% | ||
| Necrotizing enterocolitis (≥stage 2) | 2%, n=1 | 4%, n=2 | 0.642 | |
| Days to discharge, median (quartiles) | 56 (41, 74) | 40 (31, 76) | 0.0381 | |
Wilcoxon rank test;
Pearson test,
>18 hours prior to delivery
Oral colostrum priming had minimal effects on salivary immune related peptides
Salivary peptide measurements were available on unprimed (n = 35) and primed (n = 30) patients. However, no differences in maternal or neonatal variables between groups were noted on the smaller cohort (Supplemental table 1). Furthermore, not all scouted peptide sequences (Supplemental table 2) were detectable in neonatal samples. Detectable salivary peptide representation changed over time (Supplemental table 3). Specifically, we found increases in peptides corresponding to lactoferrin, immunoglobulin A, and lysozyme C and decreases in S100A7 and alpha defensin 5 at 8–9 days of life as compared to 1–2 days of life (P<0.05 by Wilcoxon rank test). No differences were detected between groups prior to OCP (DOL 1–2), which suggested the randomization process reduced starting variability (Table 2). A comparison of salivary peptide representation at DOL 8–9 in neonates that received OCP versus neonates that did not revealed a difference in the second peptide sequence for alpha defensin 1 (P<0.05 by Wilcoxon rank test). Spearman correlation between peptides is shown in Supplemental Figure 2. The percentage of saliva samples analyzed that were obtained following saline irrigation due to a lack of recoverable saliva were similar between groups [47% of OCP and 54% of control measurements, p = 0.48 by Fisher’s exact test].
Table 2.
Salivary peptide variations by group and time. Values reflect fmol detected.
| Time 1 | P-value* | Time 2 | P-value* | |||
|---|---|---|---|---|---|---|
| Peptide | No OCP | OCP | No OCP | OCP | ||
| S100a7-S1 | 22 (7–45) | 17 (8–39) | 0.68 | 20 (9–28) | 15 (8–27) | 0.5 |
| S100a7-S2 | 42 (5–169) | 35 (5–60) | 0.33 | 38 (5–88) | 18 (5–68) | 0.27 |
| S100a8-S1 | 102 (49–305) | 101 (39–313) | 0.92 | 206 (57–457) | 144 (44–308) | 0.26 |
| S100a8-S2 | 683 (109–1064) | 495 (190–854) | 0.56 | 562 (252–1037) | 511 (197–789) | 0.43 |
| S100a9-S1 | 2392 (287–3465) | 1669 (755–2540) | 0.7 | 1886 (731–4008) | 1777 (934–2473) | 0.52 |
| S100a9-S2 | 364 (67–614) | 198 (112–337) | 0.23 | 319 (114–498) | 241 (114–311) | 0.092 |
| Surfactant protein A | 0 (0–7) | 5 (0–5) | 0.83 | 5 (0–6) | 0 (0–5) | 0.4 |
| Alpha defensin 1-S1 | 1 (0–3) | 1 (0–1) | 0.32 | 1 (0–10) | 1 (0–5) | 0.41 |
| Alpha defensin 5 | 1 (1–7) | 1 (1–1) | 0.64 | 1 (0–1) | 1 (0–1) | 0.25 |
| Lactoferrin-S1 | 15 (1–40) | 11 (1–23) | 0.52 | 29 (1–91) | 57 (1–92) | 0.46 |
| Lactoferrin-S2 | 14 (8–20) | 10 (7–17) | 0.29 | 21 (12–38) | 49 (14–103) | 0.08 |
| Immunoglobulin A | 10 (10–68) | 10 (10–196) | 0.89 | 42 (10–328) | 102 (29–1373) | 0.1 |
| Lysozyme C-S1 | 1122 (559–2058) | 1257 (698–2184) | 0.89 | 1972 (1344–2617) | 1680 (836–3416) | 0.59 |
| Lysozyme C-S2 | 0 (0–20) | 0 (0–12) | 0.69 | 8 (0–23) | 15 (0–31) | 0.18 |
Median (quartiles), Time 1: 1–2 days of life; Time 2: 8–9 days of life.
Wilcoxon rank test
Oral bacterial diversity and overall composition was not altered by oral colostrum priming
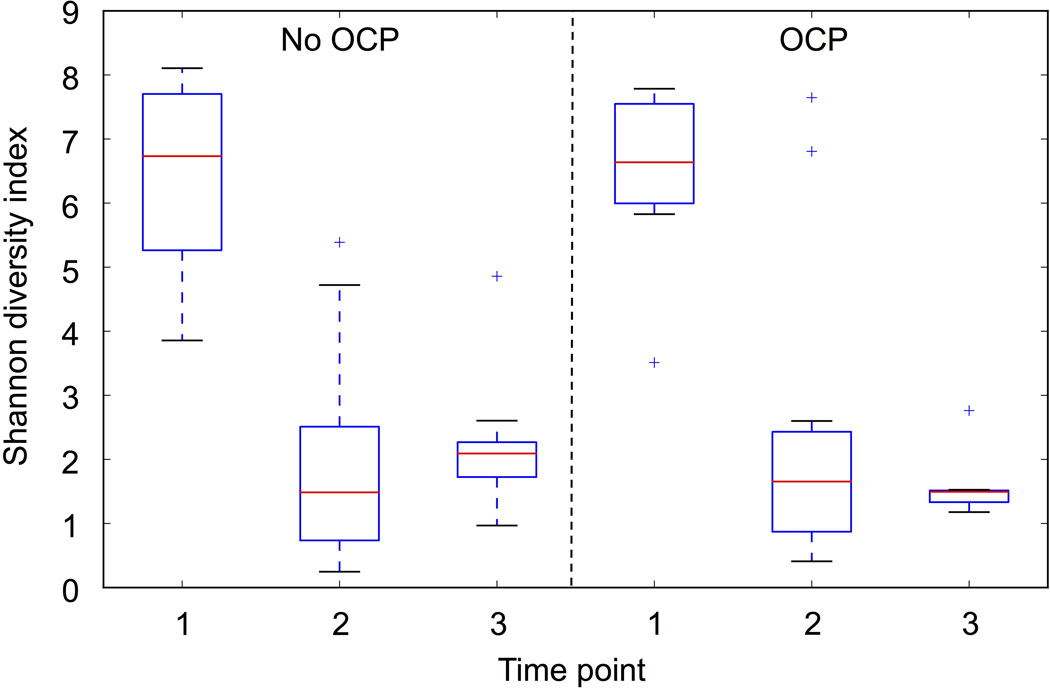
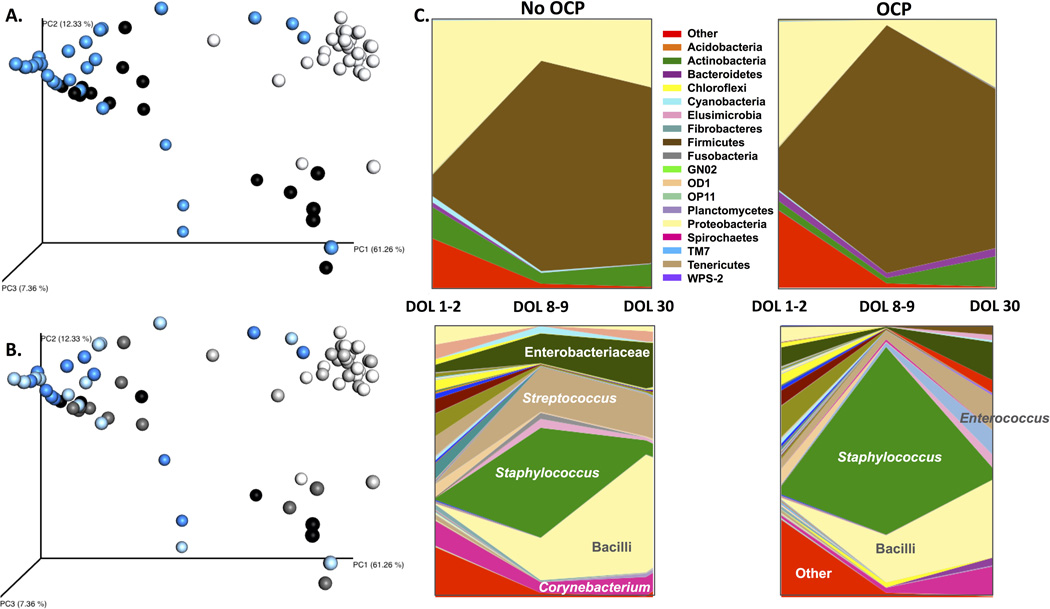
Collection of saliva samples for determination of the oral microbiota samples was added to the study protocol after more than half of the subjects had been recruited. As such, these measurements were only available on a smaller cohort of randomized patients (n = 14 unprimed, n = 14 primed). Although this is a much smaller cohort, it is the largest randomized cohort with oral microbiota determination after OCP at the time of writing (Supplemental table 4). Alpha diversity as measured by Shannon index was unchanged between OCP and no OCP groups. However, bacterial diversity significantly dropped over time (nonparametric two sample t-test with Montecarlo permutations, p<0.05) in patients across both treatment groups (Figure 2). No significant differences were observed in the overall composition of samples according to treatment (ANOSIM R= −0.007, p= 0.58); however, we found the microbiota composition was impacted by time (ANOSIM R= 0.339, p < 0.001). Principal Coordinate Analysis (PCoA) of unweighted Unifrac distance matrices were visualized as 3-D plots and demonstrated clustering of patients based on time point and not by treatment group (Figure 3A/B). Predicted functional data mirrored compositional data clustering by time point and not by treatment (Supplemental figure 3).
Figure 2. The impact of oral colostrum priming on diversity of the oral microbiota.
Shannon diversity index by group and time point. OCP-oral colostrum priming. Time points: 1 (1–2 days of life), 2 (8–9 days of life), 3 (30 days of life). Box represents 25th ad 75th percentiles, line represents median, whiskers represent maximum and minimum, and plus signs represent outliers.
Figure 3. Unweighted unifrac principal coordinate analysis (PCoA) of oral microbiota by time point and time point/ treatment group.
A. PCoA by sample time point. 1–2 days of life (n=28): white; 8–9 days of life (n=28): blue; 30 days of life (n=18): black. B. PCoA analysis by sample time point and treatment group. 1–2 days of life: white (n=14, no OCP), light grey (n=14, OCP); 8–9 days of life: light blue (n=14, no OCP), blue (n=14, OCP); 30 days of life: dark grey (n=11, no OCP), black (n=7, OCP). C. Bacterial taxa represented by group and time point. OCP-oral colostrum priming. Time points: 1 (1–2 days of life), 2 (8–9 days of life), 3 (30 days of life). Only the most relevant taxa are represented in the genus figure (bottom portion of C.).
OCP marginally impacted bacterial taxa representation
Group comparison analyses carried out between samples in treatment versus control groups showed 80 Operational Taxonomic Units (OTUs) significantly (Kruskal Wallis p< 0.05) over or under represented between treatment groups. The differences however were not significant when statistical values were corrected for multiple comparisons (FDR) (Figure 3C). Of those 80 OTUs, 69 were not assigned to known bacteria while 21 were assigned to known taxa. Of the 21 assigned taxa, 15 were decreased in the OCP group and included Streptococcus, Bifidobacterium and unclassified Enterobacteriaceae. A total of 214 OTUs were significantly altered in response to time, of which 189 were not assigned to a known bacterial species. Significant increases (Kruskal Wallis FDR corrected p < 0.05) were observed in the genera Staphylococcus and Streptococcus in both the OCP and control groups at DOL 8–9. However, abundance of Streptococcus decreased at DOL 30 only in the OCP group. Further BLAST analysis confirmed that one of the two Streptococcus OTUs differentially represented was highly similar to the human oral bacteria20. Both OCP and non-OCP groups had a temporary decrease in the relative abundance of Lactobacillus at DOL 8–9.
Discussion
Oral colostrum priming appears safe and may be associated with meaningful clinical benefits in preterm infants. Our findings contribute to our understanding of the beneficial effects through a characterization of the salivary immuno-microbial environment among patients randomized to receive OCP or no priming. Although we detected no statistically significant differences in the oral immuno-microbial environment between groups, we found a significant reduction in the length of stay (40 versus 56 days) for infants that received OCP as compared to infants that did not. This finding was not associated with differences between groups in starting time for enteral feeds, time to achieve full feeds, total days intubated, necrotizing enterocolitis, or sepsis. An unusual length of stay did not occur in our control group and thus does not explain the reduction in the length of stay we showed associated with OCP. A study of length of stay stratified by birth weight and gestational age in 30 UK NICUs found a median of 48–55 days for 1000–1250 gram infants born at 29 to 30 weeks with or without the need for respiratory support in the first 12 hours of life21. Other potential etiologies that might contribute to a difference in length of hospitalization between groups are the age at last significant apneic or bradycardic spell and the age when full oral feeding are achieved. On retrospective chart review, we did not find a difference in the number of days from the last dose of caffeine to discharge or the duration from attainment of full PO feeds to discharge. The number of days to full PO feeds was lower for infants that received OCP, but the result did not reach statistical significance. These variables would be important to consider in any future multi-center trials of OCP. Although a reduction in length of stay associated with OCP should be confirmed in larger randomized multicenter studies, OCP carries little risk and implementation should be considered.
The oral cavity is a portal for commensal and pathogenic bacteria to gain access into both the respiratory and digestive tracts22. Saliva is a proxy for the oral cavity’s microbiota23, 24 and bacteria are stable in saliva, despite constant exposure to a matrix of antimicrobial and bacterial-promoting agents25. Of note, the neonatal fecal microbiota can be significantly modified by antimicrobial exposure22, 26 and the presence of an abnormal microbiota can be associated with a host of childhood diseases states including inflammatory bowel disease, obesity, and autism27, 28. Though the gut microbiota demonstrates some degree of resiliency after antibiotic exposure, many antibiotic-induced derangements in bacterial composition may persist for long periods of time29, 3031. Importantly, we did not find statistically significant differences in the number of antibiotic days between groups of patients in our study with microbiota data. Thus, the hypothesis that oral bacterial diversity might have been altered by OCP but was attenuated by antimicrobial exposure was not supported by our findings. As shown in Table 1, the proportion of mothers who most likely would have received antibiotics prior to birth for either latency (premature rupture of membranes) or perioperative prophylaxis (C-section) was almost identical between groups. Although the possibility of pre- or peri-natal antimicrobial exposure difference between groups exists, the likelihood is low.
Composition of the oral microbiota of infants in our study correlates well with previous findings32. A recent study showed that the microbiota of infants delivered via C-section and exposed to maternal vaginal fluids resembled those of vaginally delivered infants, especially during the first week of life33. As in our study, the bacterial diversity of newborns was highest at birth (anal and oral sites) but rapidly declined. The author’s hypothesized that the drop in oral microbial diversity post-partum was the result of oral exposure to mother’s milk. Although we studied exclusively preterm infants, the group that received OCP in our study demonstrated only a marginal modification of the oral microbiota as a result of the treatment. Specifically, both groups showed increased abundances of Staphylococcus (most likely originating from the mother or caregivers’ skin) at DOL 8–9 as previously reported34, which decreased in both groups at DOL 30. Conversely, the genus Streptococcus was also significantly increased at DOL 8–9 but its abundance decreased only in the OCP group at DOL 30. Although further BLAST analysis did not allow for the full taxonomic classification of the Streptococcus OTUs altered in our study, this could be of relevance as a recent similar study with premature infants reported that one infant in their control group developed necrotizing enterocolitis at day 21 with blood culture positive for Streptococcus bovis, while a second infant had late onset group B Streptococcus sepsis at 6 weeks of life10.
Limitations and future directions
Our study is not without limitations. We restricted our investigation to infants <32 weeks gestation at birth. The protocol (volume, frequency, and duration) of oral colostrum priming used in our study was different than the regimens used in previous studies and may have contributed to differences in the findings between studies. We did not collect microbiota data on all recruited study subjects nor did we characterize the microbiota of the mother’s colostrum which could impact the oral environment35. However, once we initiated oral microbiota collection, we prospectively collected samples from patients in both groups. We also do not have environmental swabs from the neonatal intensive care unit, which may contribute to the development of the oral microbiota. However, microbial exposures are inevitable and if OCP does not modify the colonization that follows these exposures, its utility to modify infectious risk related to colonization may be limited. We feel a reduction in length of hospitalization is an important clinical and economic outcome to consider following a benign inexpensive intervention that warrants further consideration and study.
Conclusion
Although OCP did not modify the salivary peptides we examined nor overall oral bacterial diversity and composition, randomized infants that received OCP had a significantly reduced length of hospitalization, which has not previously been described. Given the safety and availability of this simple intervention, a reduction in length of stay with OCP warrants further investigation.
Supplementary Material
Acknowledgments
No form of payment was given to any author to produce the manuscript.
Funding source: Support for this work was provided by The Thrasher Fund and NIH/NIGMS [GM106143 (JLW)]
The authors wish to gratefully acknowledge the families who allowed their infants to participate in the study. We acknowledge Dr. Judy Aschner for her assistance with the study design, as well as Ms. Theresa Rogers and Ms. Amy Beller for their assistance with patient enrollment and sample collection.
Abbreviations
- OCP
oral colostrum priming
- IgA
immunoglobulin A
- VLBWs
very low birth weight infants
- DOL
day of life
- TCEP
[(tris(2-carboxyethyl)phosphine)]
- ANOSIM
analysis of similarity tests
- PCoA
Principal Coordinate Analysis
- OTUs
Operational Taxonomic Units
Footnotes
Financial disclosure statement: The authors have no financial relationships relevant to this article to disclose.
The authors have no conflicts of interest relevant to this paper.
Clinical Trial registry name and registration number: “Effect of Colostrum on Mucosal Immunity in Very Low Birth Weight (VLBWs) Premature Infants”, NCT01776268
References
- 1.Meinzen-Derr J, Poindexter B, Wrage L, Morrow AL, Stoll B, Donovan EF. Role of human milk in extremely low birth weight infants' risk of necrotizing enterocolitis or death. J Perinatol. 2009;29(1):57–62. doi: 10.1038/jp.2008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hylander MA, Strobino DM, Dhanireddy R. Human milk feedings and infection among very low birth weight infants. Pediatrics. 1998;102(3):E38. doi: 10.1542/peds.102.3.e38. [DOI] [PubMed] [Google Scholar]
- 3.Civardi E, Garofoli F, Mazzucchelli I, Angelini M, Manzoni P, Stronati M. Enteral nutrition and infections: the role of human milk. Early Hum Dev. 2014;90(Suppl 1):S57–S59. doi: 10.1016/S0378-3782(14)70019-2. [DOI] [PubMed] [Google Scholar]
- 4.Jakaitis BM, Denning PW. Human breast milk and the gastrointestinal innate immune system. Clin Perinatol. 2014;41(2):423–435. doi: 10.1016/j.clp.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Shehri SS, Knox CL, Liley HG, Cowley DM, Wright JR, Henman MG, et al. Breastmilk-Saliva Interactions Boost Innate Immunity by Regulating the Oral Microbiome in Early Infancy. PLoS One. 2015;10(9):e0135047. doi: 10.1371/journal.pone.0135047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holgerson PL, Vestman NR, Claesson R, Ohman C, Domellof M, Tanner AC, et al. Oral microbial profile discriminates breast-fed from formula-fed infants. J Pediatr Gastroenterol Nutr. 2013;56(2):127–136. doi: 10.1097/MPG.0b013e31826f2bc6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biesbroek G, Bosch AA, Wang X, Keijser BJ, Veenhoven RH, Sanders EA, et al. The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am J Respir Crit Care Med. 2014;190(3):298–308. doi: 10.1164/rccm.201401-0073OC. [DOI] [PubMed] [Google Scholar]
- 8.Maron JL, Hwang JS, Pathak S, Ruthazer R, Russell RL, Alterovitz G. Computational gene expression modeling identifies salivary biomarker analysis that predict oral feeding readiness in the newborn. J Pediatr. 2015;166(2):282–288 e285. doi: 10.1016/j.jpeds.2014.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez NA, Meier PP, Groer MW, Zeller JM, Engstrom JL, Fogg L. A pilot study to determine the safety and feasibility of oropharyngeal administration of own mother's colostrum to extremely low-birth-weight infants. Adv Neonatal Care. 10(4):206–212. doi: 10.1097/ANC.0b013e3181e94133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sohn K, Kalanetra KM, Mills DA, Underwood MA. Buccal administration of human colostrum: impact on the oral microbiota of premature infants. J Perinatol. 2016;36(2):106–111. doi: 10.1038/jp.2015.157. [DOI] [PubMed] [Google Scholar]
- 11.Lee J, Kim HS, Jung YH, Choi KY, Shin SH, Kim EK, et al. Oropharyngeal colostrum administration in extremely premature infants: an RCT. Pediatrics. 2015;135(2):e357–e366. doi: 10.1542/peds.2014-2004. [DOI] [PubMed] [Google Scholar]
- 12.Seigel JK, Smith PB, Ashley PL, Cotten CM, Herbert CC, King BA, et al. Early administration of oropharyngeal colostrum to extremely low birth weight infants. Breastfeed Med. 2013;8(6):491–495. doi: 10.1089/bfm.2013.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romano-Keeler J, Moore DJ, Wang C, Brucker RM, Fonnesbeck C, Slaughter JC, et al. Early life establishment of site-specific microbial communities in the gut. Gut microbes. 2014;5(2):192–201. doi: 10.4161/gmic.28442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards U, Rogall T, Blocker H, Emde M, Bottger EC. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17(19):7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A. 2008;105(46):17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21(3):494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manktelow B, Draper ES, Field C, Field D. Estimates of length of neonatal stay for very premature babies in the UK. Arch Dis Child Fetal Neonatal Ed. 2010;95(4):F288–F292. doi: 10.1136/adc.2009.168633. [DOI] [PubMed] [Google Scholar]
- 22.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118(2):511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 23.Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. Journal of clinical periodontology. 2003;30(7):644–654. doi: 10.1034/j.1600-051x.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- 24.Denepitiya L, Kleinberg I. A comparison of the microbial compositions of pooled human dental plaque and salivary sediment. Arch Oral Biol. 1982;27(9):739–745. doi: 10.1016/0003-9969(82)90023-1. [DOI] [PubMed] [Google Scholar]
- 25.Sakamoto M, Takeuchi Y, Umeda M, Ishikawa I, Benno Y. Application of terminal RFLP analysis to characterize oral bacterial flora in saliva of healthy subjects and patients with periodontitis. Journal of medical microbiology. 2003;52(Pt 1):79–89. doi: 10.1099/jmm.0.04991-0. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka S, Kobayashi T, Songjinda P, Tateyama A, Tsubouchi M, Kiyohara C, et al. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol Med Microbiol. 2009;56(1):80–87. doi: 10.1111/j.1574-695X.2009.00553.x. [DOI] [PubMed] [Google Scholar]
- 27.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 28.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155(7):1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De La Cochetiere MF, Durand T, Lepage P, Bourreille A, Galmiche JP, Dore J. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J Clin Microbiol. 2005;43(11):5588–5592. doi: 10.1128/JCM.43.11.5588-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1(1):56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 31.Gronlund MM, Lehtonen OP, Eerola E, Kero P. Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J Pediatr Gastroenterol Nutr. 1999;28(1):19–25. doi: 10.1097/00005176-199901000-00007. [DOI] [PubMed] [Google Scholar]
- 32.Hendricks-Munoz KD, Xu J, Parikh HI, Xu P, Fettweis JM, Kim Y, et al. Skin-to-Skin Care and the Development of the Preterm Infant Oral Microbiome. Am J Perinatol. 2015;32(13):1205–1216. doi: 10.1055/s-0035-1552941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med. 2016 doi: 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sampaio-Maia B, Monteiro-Silva F. Acquisition and maturation of oral microbiome throughout childhood: An update. Dent Res J (Isfahan) 2014;11(3):291–301. [PMC free article] [PubMed] [Google Scholar]
- 35.Cacho N, Neu J. Manipulation of the intestinal microbiome in newborn infants. Adv Nutr. 2014;5(1):114–118. doi: 10.3945/an.113.004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.