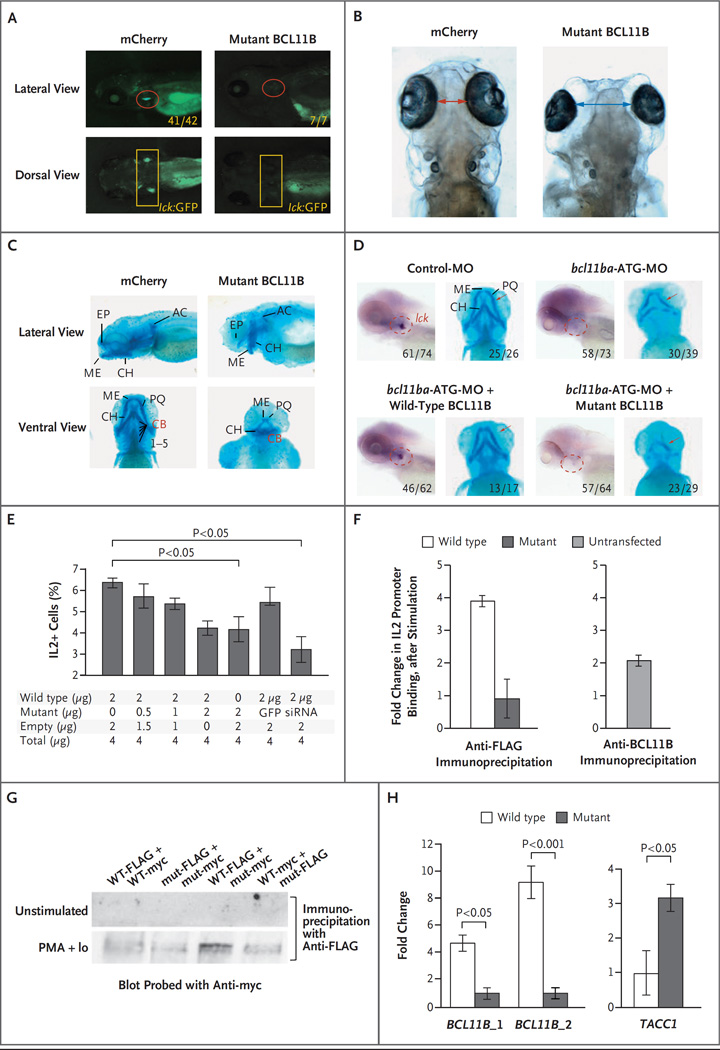
Figure 3 (facing page). Dominant Negative Effect of Mutant BCL11B in Zebrafish and Jurkat T Cells.
Panel A shows T-cell development in the zebrafish thymus at 5 days after fertilization after either human mutant BCL11B or mCherry as a control was overexpressed by injection of mRNA (25 pg) into transgenic lck:GFP embryos. Panels B and C show increased intraocular distance (arrows) and disruption of cartilaginous cranial structures 5 days after fertilization in zebrafish overexpressing human p.N441K BCL11B (as in Panel A). The intraocular distance increased from a mean of 1.0 mm to 2.5 mm (five embryos, P<0.001). Panel D shows the ability of intact but not mutant human BCL11B to rescue the arrest in T-cell development and correct the craniofacial abnormalities caused by bcl11ba knockdown. Rescue of T-cell development was evaluated after heat-inducible reexpression (30 hours after fertilization) and whole-mount in situ hybridization (WISH) analysis with an lck probe at 5 days after fertilization to identify thymocytes (within dashed red outlines in the lateral view). The restoration of craniofacial cartilage structure (red arrows) was assessed at 5 days after fertilization with the use of Alcian blue staining. Numbers indicate the proportions of embryos that had the depicted phenotypes. Panel E shows the production of interleukin-2 by Jurkat cells that were stimulated with phorbol myristate acetate (PMA) and ionomycin. The cells were transfected with a constant amount of vector DNA, with increasing proportions of p.N441K BCL11B, GFP vector, or BCL11B siRNA. I bars indicate standard deviations. Panel F shows chromatin immunoprecipitation (ChIP) after stimulation, with PMA and ionomycin, of Jurkat cells (left panel) transfected with FLAG-tagged wild-type or p.N441K BCL11B, and untransfected Jurkat cells (right panel). The fold change in DNA was calculated as the ratio of output DNA to input DNA in cells stimulated with PMA and ionomycin relative to the ratio in unstimulated cells. ChIP in untransfected cells was performed with anti-BCL11B antibody, as a control. Panel G shows the ability of BCL11B to heterodimerize, assessed by transfection of distinctly tagged wild-type (WT) and mutant (mut) BCL11B into Jurkat cells and coprecipitation with the indicated anti-tag antibodies. Extracts were produced from unstimulated Jurkat cells and from cells stimulated with PMA and ionomycin (PMA+Io). Panel H shows verification, by quantitative polymerase chain reaction (PCR), of BCL11B binding sites in genomic loci detected by ChIP-seq analysis of human cord-blood progenitors that were expanded in vitro. DNA bound by FLAG-tagged wild-type or mutant BCL11B was isolated by coprecipitation with anti-FLAG antibody. Fold changes were calculated as the relative quantity of the DNA binding site of interest (BCL11B_1, BCL11B_2, or TACC1), determined by quantitative PCR in ChIP-seq samples from human cord-blood progenitors transduced with FLAG-tagged wild-type BCL11B relative to that in samples transduced with FLAG-tagged mutant BCL11B. Two canonical BCL11B binding sites within the BCL11B locus were bound more extensively (one site 5 times more extensively and one site 10 times more extensively) than were the sites in the p.N441K mutant (left panel). Conversely, a novel TACC1 site (isoform 1, NM_006283.2) was found to have greater binding by the p.N441K mutant than by wild-type BCL11B.