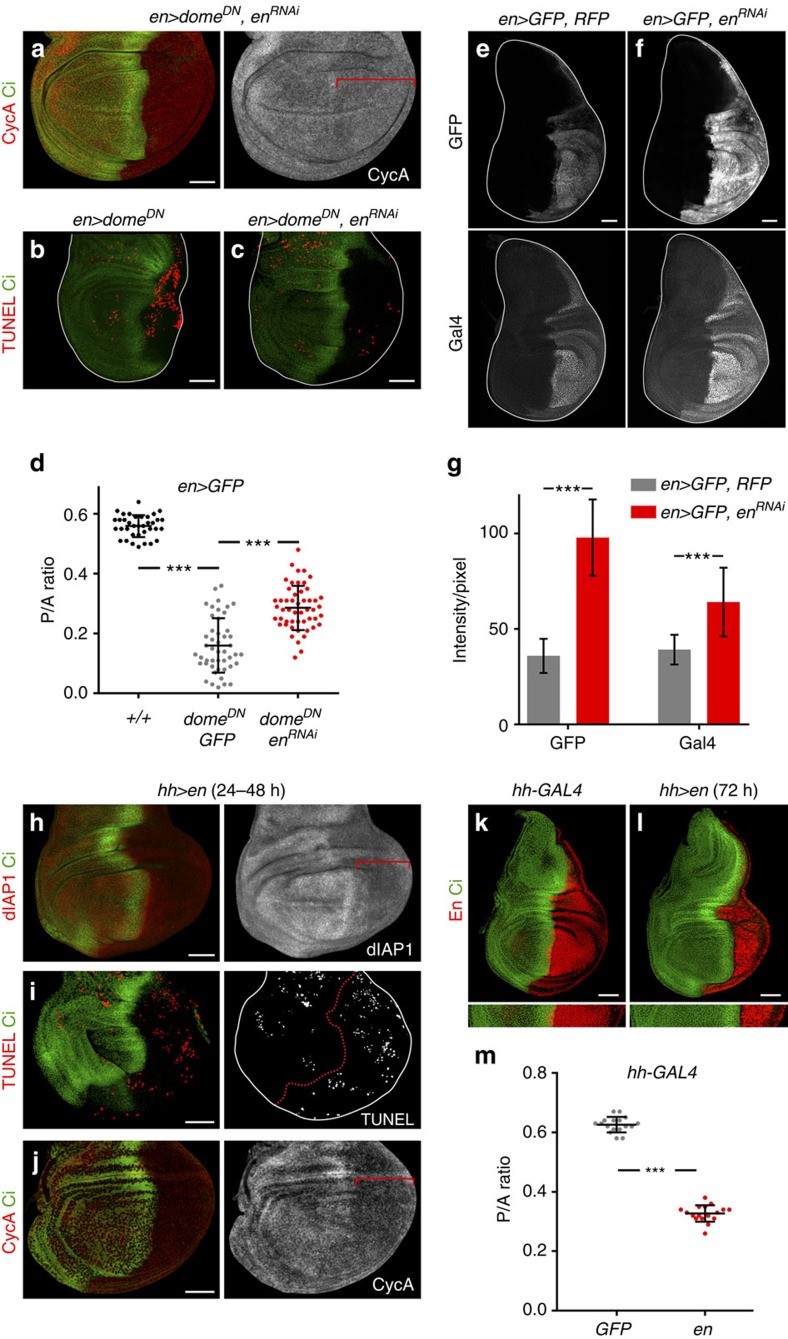
Figure 6. JAK/STAT counteracts En activity on survival and cycling of P cells.
(a–c) Late third instar wing primordia from larvae of the indicated genotypes labelled to visualize Cyclin A (CycA, red or white, a), apoptotic cells by TUNEL staining (red, b,c) and Ci (green). (e,f) Late third instar wing primordia from larvae of the indicated genotypes labelled to visualize GFP or Gal4 (white). (g) Bar graph showing the average signal intensity/pixel of GFP and Gal4 protein levels of the indicated genotypes. For en>GFP, RFP, GFP intensity=35.93±8.93; Gal4 intensity=39.14±7.79 (n=18). For en>GFP, enRNAi-VDRC, GFP intensity=97.86±19.88; Gal4 intensity=64.13±18.27 (n=23). Error bars represent s.d. P values: Student's t-test, ***P<0.001. (h–l) Late third instar wing primordia from individuals overexpressing En (h–j,l) or GFP (k) in the P compartment for 24–48 h (h–j), or 72 h (k,l), and labelled to visualize dIAP1 protein (red or white, h), apoptotic cells by TUNEL staining (red or white, i), CycA (red or white, j), En (red, k,l) and Ci (green) protein expression. Red brackets in a,h,j mark the domain of transgene expression; wing disc contours are marked in b,c,e,f,i by a white line; red dashed line in i marks the AP boundary. Scale bars, 50 μm. (d,m) Scatter plots showing the P/A ratio of wing primordia expressing the indicated transgenes under the control of the en-gal4 (d) or hh-gal4 (m) drivers. In d, P/A ratios from left to right: 0.56±0.04 (n=36); 0.16±0.09 (n=47); 0.29±0.07 (n=51). In m, P/A ratios from left to right: 0.63±0.03 (n=17); 0.33±0.03 (n=17). Error bars (vertical lines) represent s.d. and long horizontal lines mark average values. P values: Student's t-test, ***P<0.001.