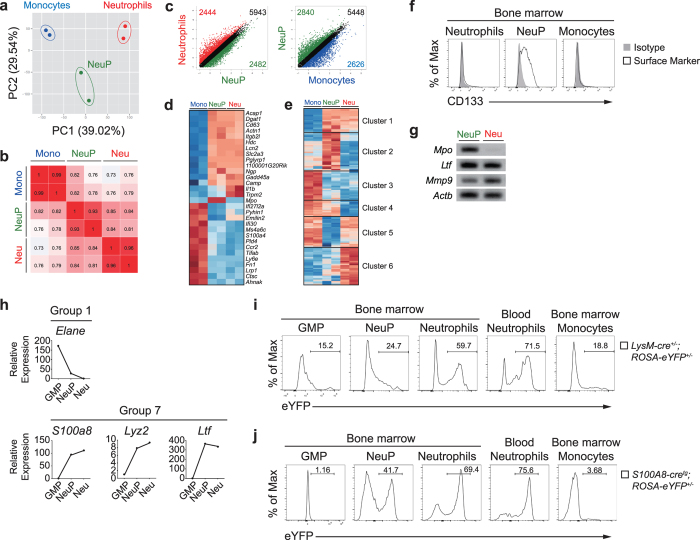
Figure 4. Transcriptome analysis of NeuPs.
(a) PCA analysis of gene expression profiles of monocytes, NeuPs, and neutrophils. Numbers in parenthesis indicate the percentage of the total variance represented by the axis. (b) Correlation matrix of monocytes, NeuPs, and neutrophils based on all available genes. Numbers in plots indicate correlation coefficient values. (c) Diversity of gene expression in monocytes, NeuPs, and neutrophils. Numbers in plots indicate probes with a minimum change of 2-fold in expression. Cell types are color-coded. (d) Heat map and hierarchical clustering of mRNA transcripts of DEGs determined by Cuffdiff (FDR < 0.05). (e) Heat map of genes encoding transcription factors whose expression showed more than a 3-fold change between cell types and less than a 2-fold difference between replicates. Pearson distance is used as a distance function. Genes were clustered into six groups and are listed in Table 1. (f) Expression level of CD133, novel candidate for NeuP specific surface marker, on NeuPs, neutrophils and monocytes were detected with FACS. (g) RT-PCR of genes for granule markers in NeuPs and neutrophils. This is a processed and cropped version. The original full-length gel image is presented in the Supplementary Figure S9. (h) Expression of neutrophil lineage genes (Elane, S100a8, Lyz2, and Ltf) among GMPs, NeuPs, and neutrophils. Relative expression compared to neutrophils is shown. (i-j) EYFP labeling of GMPs, NeuPs, neutrophils, and monocytes based on Lyz2 expression by using Lyz2-cre; ROSA-eYFP mice (i) or on S100a8 expression by using S100A8-cre; ROSA-eYFP mice (j). Percentage of eYFP-expressing cells, determined in comparison to WT mice, is shown. Data are representative of two independent experiments.