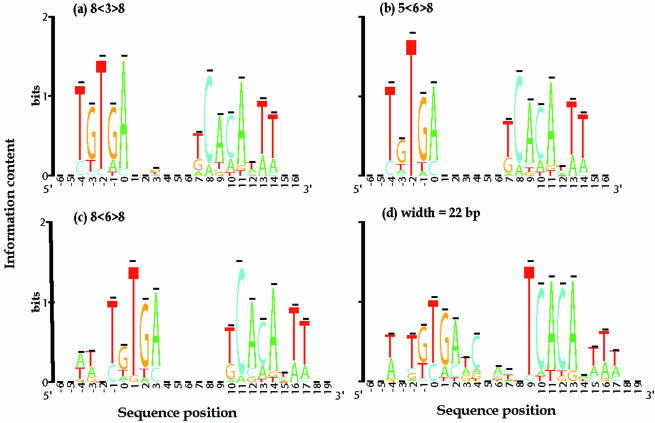
Figure 6.
Sequence logos for one-block and bipartite models of CRP-binding sites. (a) Bipartite pattern 8<3>8, dominant spacer length is 3 bp; information in left half site is dominated by five positions (−4 to 0) with positions 1, 2, and 3 considered noise; total information for left site is Rseq(left) = 6.03 bits per 8 bp and for the right-site Rseq(right) = 6.62 bits per 8 bp; after removing these noise signals the total information for both half sites increased and the model was reduced to that seen in panel (b). (b) Bipartite pattern 5<6>8, dominant spacer lengths are 6 bp; total information for left site is Rseq(left) = 5.65 bits per 5 bp and for right-site Rseq(right) = 7.02 bits per 8 bp. (c) 5<6>8 extended to 8<6>8 with Rseq(left) = 6.04 bits and Rseq(right) = 7.19 bits. (d) Motif width of one-block model is 22 bp as demonstrated in DNA footprint studies; the total information for this model is Rseq(one-block) = 10.42 bits per 22 bp. Thus, the bipartite model contains >2 bits per site more than the one-block model and it is obvious from the distribution of information in the logos that the pattern can be subdivided into two half sites.