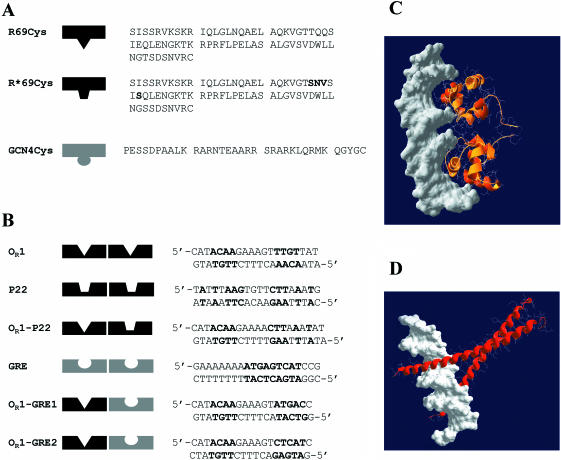
Figure 1.
(A) Amino acid sequences of R69Cys and R*69Cys. R69Cys and R*69Cys differ in the sequence of the recognition helix (the mutated residues in R*69Cys are shown in bold) (B) Sequences of various synthetic oligonucleotides used in the study. The nucleotides shown in bold represent the consensus sequence of the specific binding sites. OR1 corresponds to the OR1 site of the operator sequence of 434 repressor. P22 is the sequence of the operator OR1 site of phage P22 repressor c2. GRE contains the specific operator sequence of GCN4 repressor. OR1–P22 represents a hybrid DNA with one 434-specific and one P22-specific half-sites. OR1–GRE1 and OR1–GRE2 represent hybrid DNAs with one 434-specific and one GCN4-specific half-sites: they differ in the orientation of the GRE half-site. (C) Computer model of two R69Cys monomers in complex with DNA. The coordinates of residues 1–69 were taken from the NMR structure of R69 in solution (Brookhaven PDB 1PRA) and superimposed (1–63 residues) on the peptide backbone of the resolved X-ray R69/OR2 complex (1RPE). The model was constructed using the program Swiss-PdbViewer v3.6b3. (D) Computer model of two GCN4 monomers in complex with DNA (Brookhaven PDB 1YSA).