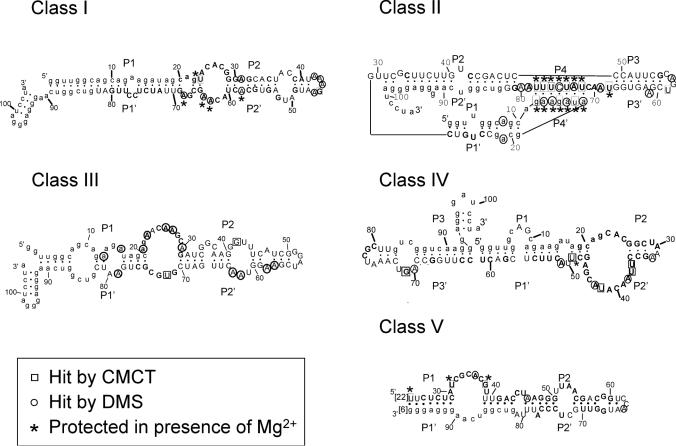
Figure 5.
Secondary structure of the five ATP-binding motifs. Secondary structure models of the five aptamer motifs as defined by structure probing, sequence comparisons and energy minimization (42). The sequences shown are those of clone C14 (class I), C20 (class II), C27 (class III), C15 (class IV) and C25 (class V). Squares and circles indicate nucleotides hit by CMCT and DMS, respectively. A–U and G–U base pairs at the ends of helices have been implemented even if one of the residues is hit by a modifying agent. Pn's designate the paired segments. The sequences of conserved regions (see Figure 6) are shown in bold, the variability within theses sequences is indicated in Figure 7. Primer sequences were omitted from class V aptamers secondary structure model because they are not involved in any potential pairing. The number of nucleotides omitted is indicated in between brackets.