Fig. 1.
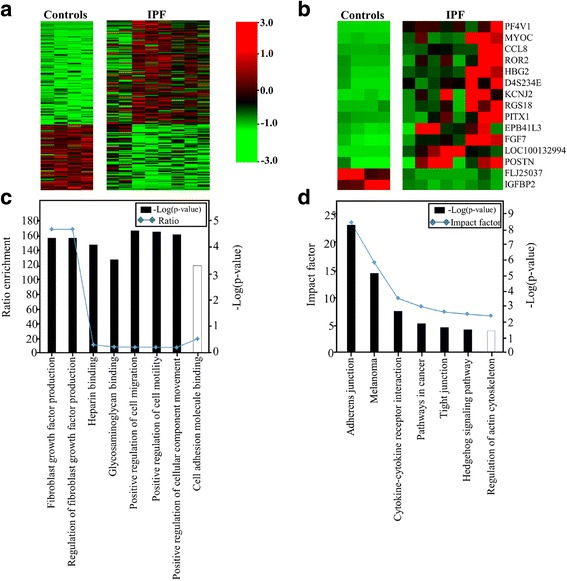
Gene expression profiles of fibroblasts derived from the lung tissues of 8 patients with IPF and 4 controls. a A heat map of 178 genes differentially expressed between the two groups (p-value <0.05 and absolute fold-change ratio >2 by t-test and TNoM). The maximum value (red) of each gene was set to 3, the minimum value to −3, and the remaining values were linearly fitted in the range. b A heat map of top 15 genes differentially expressed between the two groups (p-value < 0.05 by t-test and TNoM, absolute fold-change ratio >10). c The top 8 significantly perturbed Gene Ontology nodes in the IPF patients versus the controls. Left, statistical significance of the perturbation as determined by a gene set test; right, ratio of enrichment. The significance of differences between the dataset and the canonical pathway was measured as a ratio. Solid and open bars represent upregulation and downregulation, respectively. d Biological pathway analysis of differentially expressed gene sets related to IPF (corrected gamma p-value < 0.05). P-values and impact factors are plotted on the left and right axes, respectively
