Fig. 1.
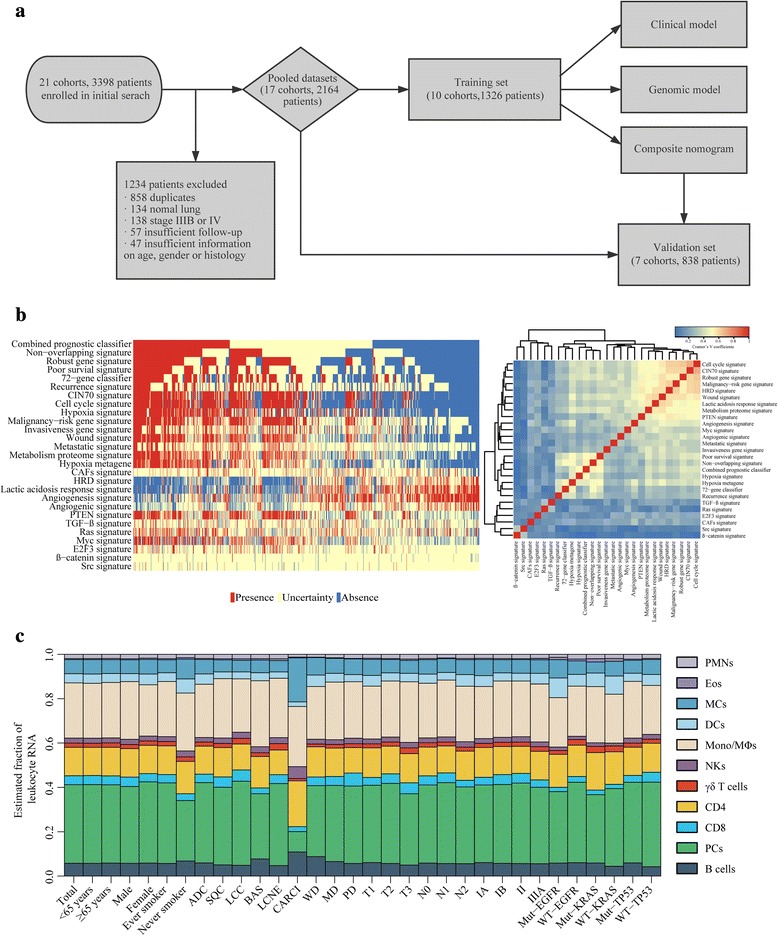
Genomic landscape of operable NSCLC based on gene expression profiling. a Flow chart of the study design. b Concordance of signature-based prediction result in the training set. Left panel: Each column represents the prediction of each individual sample. Red, blue and pale yellow bars indicate presence, absence and uncertainty prediction of the corresponding signature, respectively. Right panel: Heatmap of Cramer’s V coefficient showing correlation between these published signatures. c Consistency of estimated mRNA fractions of 11 tumor-associated Leukocyte (TAL) subsets, as calculated by CIBERSORT, within and across clinical subgroups of NSCLC in the training set
