Abstract
We discuss the generic time behavior of reaction-diffusion processes capable of modeling various types of biological transport processes, such as ligand migration in proteins and gating fluctuations in ion channel proteins. The main observable in these two cases, the fraction of unbound ligands and the probability of finding the channel in the closed state, respectively, exhibits an algebraic t-1/2 decay at intermediate times, followed by an exponential cutoff. We provide a simple framework for understanding these observations and explain their ubiquity by showing that these qualitative results are independent of space dimension. We also derive an experimental criterion to distinguish between a one-dimensional process and one whose effective dimension is higher.
Full text
PDF
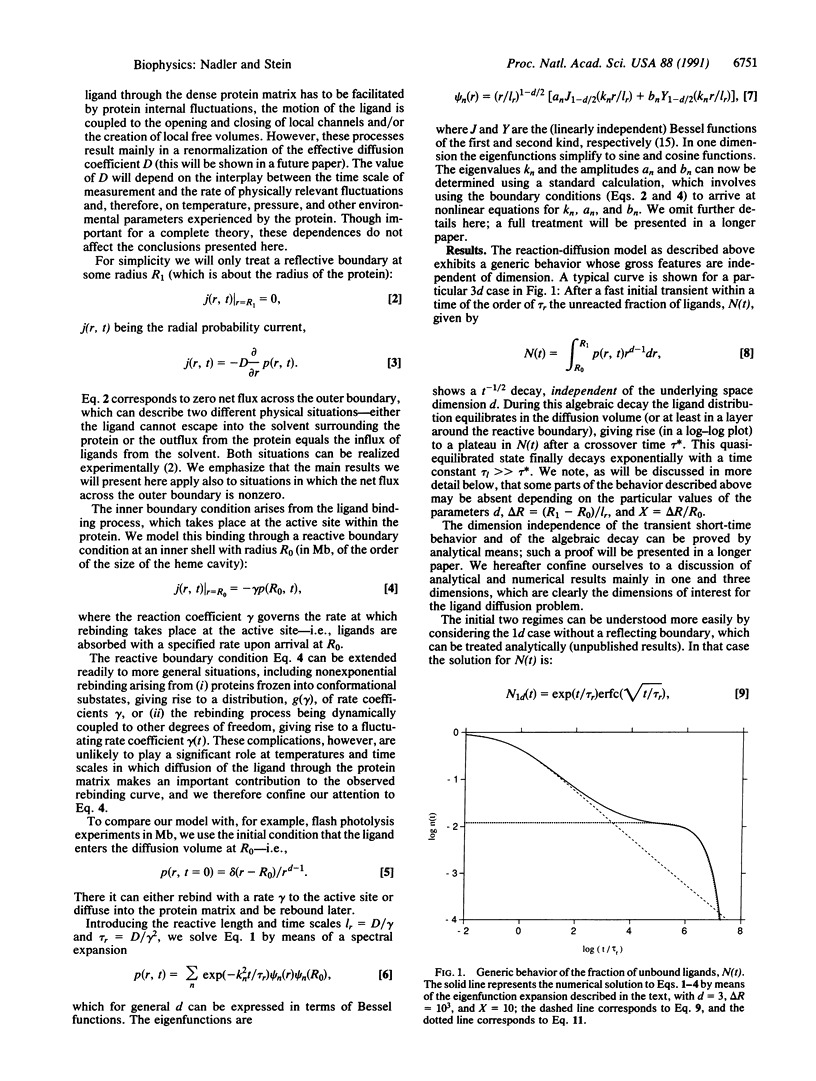
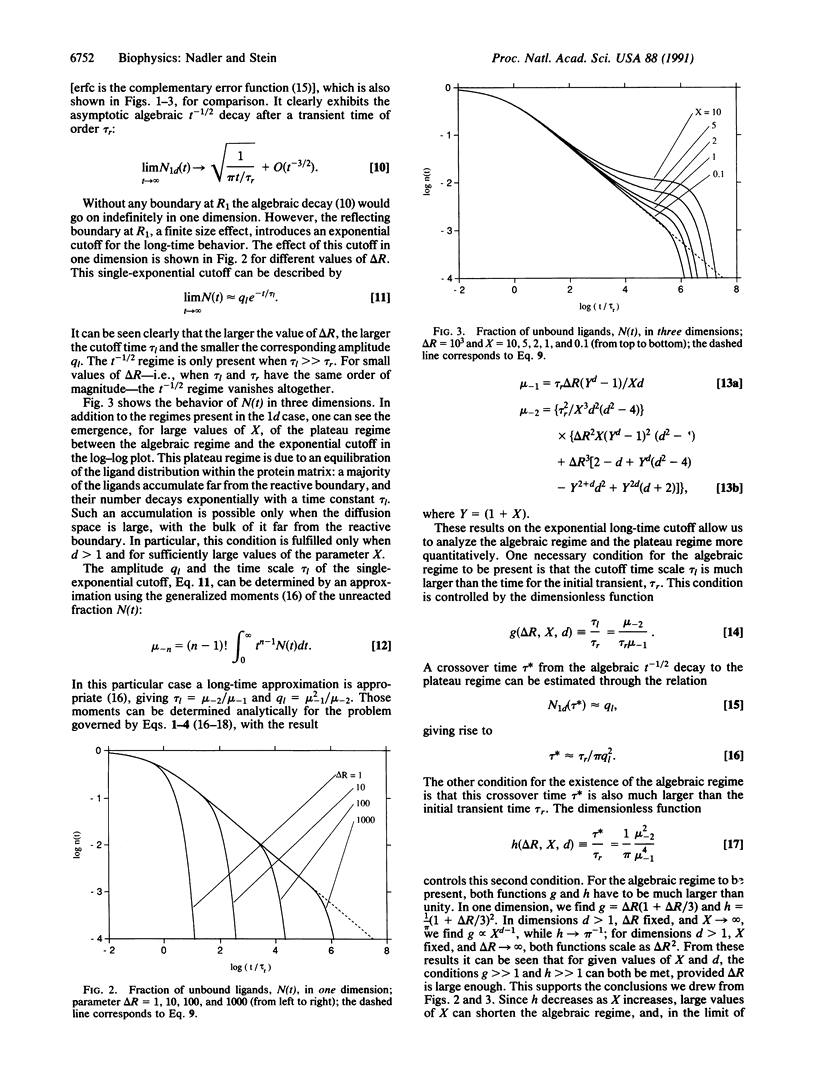


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberding N., Chan S. S., Eisenstein L., Frauenfelder H., Good D., Gunsalus I. C., Nordlund T. M., Perutz M. F., Reynolds A. H., Sorensen L. B. Binding of carbon monoxide to isolated hemoglobin chains. Biochemistry. 1978 Jan 10;17(1):43–51. doi: 10.1021/bi00594a007. [DOI] [PubMed] [Google Scholar]
- Ansari A., Berendzen J., Bowne S. F., Frauenfelder H., Iben I. E., Sauke T. B., Shyamsunder E., Young R. D. Protein states and proteinquakes. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5000–5004. doi: 10.1073/pnas.82.15.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A., Berendzen J., Braunstein D., Cowen B. R., Frauenfelder H., Hong M. K., Iben I. E., Johnson J. B., Ormos P., Sauke T. B. Rebinding and relaxation in the myoglobin pocket. Biophys Chem. 1987 May 9;26(2-3):337–355. doi: 10.1016/0301-4622(87)80034-0. [DOI] [PubMed] [Google Scholar]
- Ansari A., DiIorio E. E., Dlott D. D., Frauenfelder H., Iben I. E., Langer P., Roder H., Sauke T. B., Shyamsunder E. Ligand binding to heme proteins: relevance of low-temperature data. Biochemistry. 1986 Jun 3;25(11):3139–3146. doi: 10.1021/bi00359a011. [DOI] [PubMed] [Google Scholar]
- Austin R. H., Beeson K. W., Eisenstein L., Frauenfelder H., Gunsalus I. C. Dynamics of ligand binding to myoglobin. Biochemistry. 1975 Dec 2;14(24):5355–5373. doi: 10.1021/bi00695a021. [DOI] [PubMed] [Google Scholar]
- Beece D., Eisenstein L., Frauenfelder H., Good D., Marden M. C., Reinisch L., Reynolds A. H., Sorensen L. B., Yue K. T. Solvent viscosity and protein dynamics. Biochemistry. 1980 Nov 11;19(23):5147–5157. doi: 10.1021/bi00564a001. [DOI] [PubMed] [Google Scholar]
- Case D. A., Karplus M. Dynamics of ligand binding to heme proteins. J Mol Biol. 1979 Aug 15;132(3):343–368. doi: 10.1016/0022-2836(79)90265-1. [DOI] [PubMed] [Google Scholar]
- Condat C. A., Jäckle J. Closed-time distribution of ionic channels. Analytical solution to a one-dimensional defect-diffusion model. Biophys J. 1989 May;55(5):915–925. doi: 10.1016/S0006-3495(89)82890-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condat CA. Defect diffusion and closed-time distributions for ionic channels in cell membranes. Phys Rev A Gen Phys. 1989 Feb 15;39(4):2112–2125. doi: 10.1103/physreva.39.2112. [DOI] [PubMed] [Google Scholar]
- Doster W., Beece D., Bowne S. F., DiIorio E. E., Eisenstein L., Frauenfelder H., Reinisch L., Shyamsunder E., Winterhalter K. H., Yue K. T. Control and pH dependence of ligand binding to heme proteins. Biochemistry. 1982 Sep 28;21(20):4831–4839. doi: 10.1021/bi00263a001. [DOI] [PubMed] [Google Scholar]
- Doster W., Schirmacher W., Settles M. Percolation model of ionic channel dynamics. Biophys J. 1990 Mar;57(3):681–684. doi: 10.1016/S0006-3495(90)82588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M., McCammon J. A. The internal dynamics of globular proteins. CRC Crit Rev Biochem. 1981;9(4):293–349. doi: 10.3109/10409238109105437. [DOI] [PubMed] [Google Scholar]
- Korn S. J., Horn R. Statistical discrimination of fractal and Markov models of single-channel gating. Biophys J. 1988 Nov;54(5):871–877. doi: 10.1016/S0006-3495(88)83023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Läuger P. Internal motions in proteins and gating kinetics of ionic channels. Biophys J. 1988 Jun;53(6):877–884. doi: 10.1016/S0006-3495(88)83168-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus O. B., Weiss D. S., Spivak C. E., Blatz A. L., Magleby K. L. Fractal models are inadequate for the kinetics of four different ion channels. Biophys J. 1988 Nov;54(5):859–870. doi: 10.1016/S0006-3495(88)83022-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millhauser G. L., Oswald R. E. A reevaluation of the mathematical models for simulating single-channel and whole-cell ionic currents. Synapse. 1988;2(1):97–103. doi: 10.1002/syn.890020113. [DOI] [PubMed] [Google Scholar]
- Millhauser G. L. Reptation theory of ion channel gating. Biophys J. 1990 Apr;57(4):857–864. doi: 10.1016/S0006-3495(90)82605-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millhauser G. L., Salpeter E. E., Oswald R. E. Diffusion models of ion-channel gating and the origin of power-law distributions from single-channel recording. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1503–1507. doi: 10.1073/pnas.85.5.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millhauser G. L., Salpeter E. E., Oswald R. E. Rate-amplitude correlation from single-channel records. A hidden structure in ion channel gating kinetics? Biophys J. 1988 Dec;54(6):1165–1168. doi: 10.1016/S0006-3495(88)83051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]


