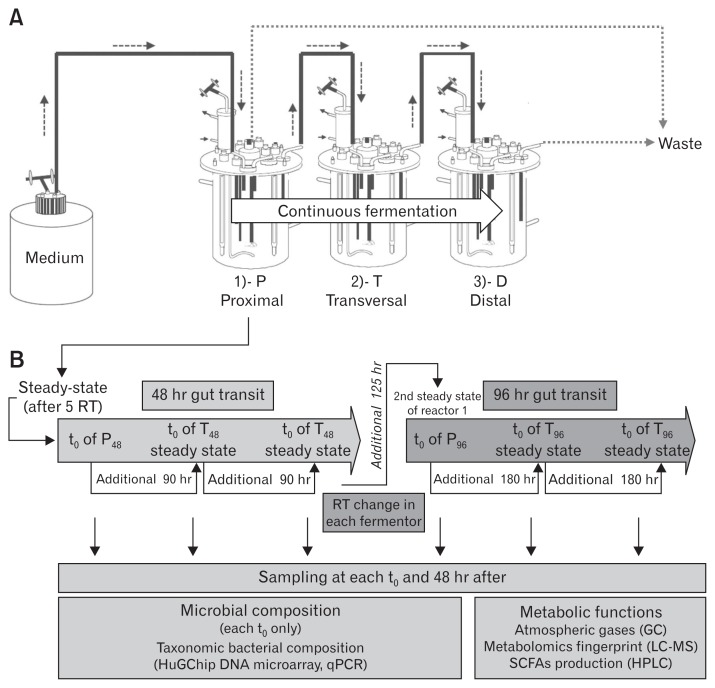
Figure 1.
The 3-stage Environmental Control System for Intestinal Microbiota (3S-ECSIM) and overall design of the experiment. (A) The 3S-ECSIM system is depicted with its 3 reactors: proximal, transversal, and distal (P, T, and D), respectively, simulating the physicochemical parameters of the proximal, transversal, and distal parts of the colon. Each reactor is initially inoculated by the same regenerated fecal inoculum. The first one is fed with a growth medium simulating nutrients entering the proximal colon in a continuous fermentation process, using a retention time (RT) fitting with first a normal transit time (48 hours) and next a slow one (96 hours) (see Table 1 for parameters). Resulting medium from the P reactor is continuously used to feed the second reactor (T), and next the third reactor (D) in a similar process. Volumes of reaction are maintained by retrieving excess of medium to fit also with RT (reactors P and D, see Table 1). (B) After at least 5 RT of continuous feeding, the gut microbiota is considered to be in a steady-state (modeled at 96% stability) and the resulting metabolized medium considered to be stable. This time corresponds to t0 in each reactor (see additional data, at time t0 and 48 hours after, for metabolic comparisons at steady state). For a 48-hour gut transit simulation (orange, left part), the resulting medium from the P reactor is used to feed the second reactor during 90 hours more (in case of a gut normal transit time of 48 hours, Table 1) in order to obtain 5 RT in the T reactor. The same principles are applied for the third reactor (D48) and the medium resulting from reactor T48. The RT is next changed in each compartment in order to simulate a slow transit time (96 hours all over the gut, blue, right part) and similar principles are applied (see Table 1 for parameters). At each t0 and 48 hours after, samplings are done and analyses performed (in grey boxes, lower part). This encompasses the determination of the microbial composition by the Human Gut Chip (HuGChip), a DNA phylogenetic microarray detecting 16S bacterial sequences,20 and its metabolic behavior by measuring the fermentative atmospheric gases, the production of short chain fatty acids (SCFAs) and by determining a high resolution metabolite fingerprint.