Abstract
We have developed a software package called Osprey for the calculation of optimal oligonucleotides for DNA sequencing and the creation of microarrays based on either PCR-products or directly spotted oligomers. It incorporates a novel use of position-specific scoring matrices, for the sensitive and specific identification of secondary binding sites anywhere in the target sequence. Using accelerated hardware is faster and more efficient than the traditional pairwise alignments used in most oligo-design software. Osprey consists of a module for target site selection based on user input, novel utilities for dealing with problematic sequences such as repeats, and a common code base for the identification of optimal oligonucleotides from the target list. Overall, these improvements provide a program that, without major increases in run time, reflects current DNA thermodynamics models, improves specificity and reduces the user's data preprocessing and parameterization requirements. Using a TimeLogic™ hardware accelerator, we report up to 50-fold reduction in search time versus a linear search strategy. Target sites may be derived from computer analysis of DNA sequence assemblies in the case of sequencing efforts, or genome or EST analysis in the case of microarray development in both prokaryotes and eukaryotes.
INTRODUCTION
Oligonucleotides (oligos) have many applications in molecular biology, and there are many programs that can be used for their calculation. While it is still fairly common to design oligo PCR-primers manually using the so-called Wallace Rule: G/C = 4°C, A/T = 2°C, summed for melting temperature (1); more accurate formulae exist that closely model the thermodynamics of nucleotide binding. The need to calculate large numbers of primers for genomic sequencing and the growing use of microarrays have led to the development of increasingly sophisticated algorithms to improve the automation of oligo design. These algorithms have been the subject of several recent papers (2,3).
Non-target binding can cause sequencing reactions to be unusable, and give false mRNA expression level readings in microarrays. Eliminating this is diversely implemented. Oligodb (4) filters out low-complexity regions using dustn (http://www.ncbi.nlm.nih.gov/IEB/ToolBox/) without checking if the sequences are repeated. PROBEWIZ (5) explicitly disables filtering, while other system manuals do not document this aspect of the computation. Most programs use a simple BLAST (6) search to filter secondary binding based on percent mismatch, but this method has disadvantages; small sequence stretches with evenly spaced mismatches may not be found due to the heuristic nature of BLAST. Even if these methods could find all matches, the Sarani documentation (http://www.strandgenomics.com) shows that the duplex melting temperature of two 20 base targets against the same oligo sequence can differ by 20°C when both targets have only two mismatches. Also, high GC regions bind with much higher energy than low GC regions of similar length. The number of pairwise matches is therefore not necessarily a good measure of melting thermodynamics. OligoArray (7) compensates for mismatches with iterative rounds of BLAST searches with decreasing mismatch tolerance. Sarani compensates with a specialized BLAST-like search where the extension of hashed high-scoring pairs is based on the thermodynamic criterion of the alignment.
SantaLucia's ‘unified’ free energy parameters model (8) was derived from the unification of previously described nearest neighbor (NN) methods, and is generally considered the best model yet of DNA binding thermodynamics for melting temperature and duplex stability. The NN model assumes that summing the interaction energy of adjacent nucleic acids on a strand is the best predictor of the whole duplex's stability. SantaLucia's formula is used by the high-throughput commercial packages Sarani and Array Designer 2 (http://www.premierbiosoft.com), as well as by the interactive GeneFisher (9). Other programs are generally based on older formulae (10,11), most often because they incorporate Primer3 (12) as a software component. Primer3 is popular because its predictions have proven very useful in practice. But SantaLucia observes that the melting temperature [generally considered the point at which half the duplexes are annealed (13)] in the Breslauer model, on which Primer3 is based, has a standard deviation of 6°C for the unified model's reference oligo data set. This can cause complications if a narrow melting temperature range is desired for a large data set such as a microarray: a 10°C window grows to 22°C.
SantaLucia's HyTher™ Web server (http://ozone2.chem.wayne.edu) includes other data such as those for internal mismatches (14–18) and dangling end mismatches (19). Dangling ends, which almost always occur unless the oligo and target have the same length, can contribute more to double-strand stability than an A–T neighbor pairing. None of the widely available primer design applications describe using these extra data.
Osprey was built to calculate sequencing primers for the Sulfolobus solfataricus P2 genome (20). To save labor and primer costs, we required software that took all the standard design parameters into account, and designed a minimal set of primers directly from assembly data with little human intervention. Later projects required its adaptation to design primers for PCR-based microarrays (21) and directly spotted microarrays (70mer oligos). We describe here oligo selection in Osprey, focusing on duplex formation efficiency and the commonalities of the computation among different oligo design tasks. We highlight automated techniques that provide higher quality oligos with less human intervention, and we introduce the novel use of position-specific scoring matrices (PSSMs) to encode the free energy model, improving the specificity and sensitivity of oligo secondary binding searches.
METHODS
Osprey implementation
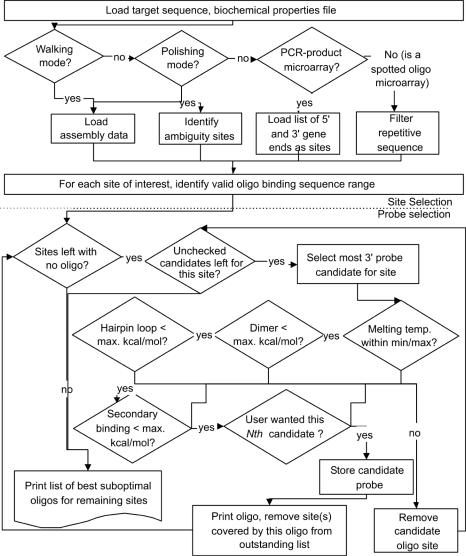
As illustrated in Figure 1, Osprey has two main stages: (i) a set of all possible oligos is created based on the user-selected search mode, target sequence range and oligo size; and (ii) oligo candidates pass through a series of fitness exclusion tests. Osprey is designed to parallelize processes wherever feasible to facilitate large-scale oligo selection. We will describe each fitness module: melting temperature, dimer formation, hairpin formation and secondary binding. A configuration file contains biophysical parameters for these tests, including temperature and energy cutoff values, DNA and salt concentrations, and sequencing performance expectations. Command line arguments specify the files containing the sequence, assembly information and gene locations. Users should override the provided default values using the command line or Web interface to match the experimental conditions and availability of PSSM search accelerators discussed later in this text.
Figure 1.
Workflow diagram of oligo selection in Osprey. Sequences, assembly information and default parameters are read from disk. Many parameters and modes can be overridden on the command line. Note that the probe selection part of the processing is common to all oligo design modes. Also, the probe selection test cascade can be run concurrently for many sites.
Selection of the oligonucleotide location
Osprey has varying search strategies for four different types of oligos: (i) primers for DNA sequencing that can be used to link contigs (linking primers), (ii) primers for DNA sequencing that can be used for the disambiguation or to double-strand DNA sequence assemblies (polishing primers), (iii) primers for PCR, including those used for microarrays and (iv) oligos for the direct spotting of microarrays (e.g. glass slides with 70mer oligos). All modes require users to provide information about the biochemical environment via a configuration file in order to properly use the thermodynamic model.
DNA sequencing primers
Osprey can be controlled using a contig information file from the Staden sequence assembly package (22). Data generated using the ‘Show relationships’ option in gap4 is saved to disk as plain text, to which user can add simple control information. The program calculates default primers only for those contiguous sequences (contigs) that are not marked IGNORE on the left-hand side of the line denoting contig neighbors. In walking mode, NO 5PRIME on that line location will cause the 5′ primer not to be calculated. Other valid directives are NO 3PRIME, NTH 3PRIME and NTH 5PRIME, where N chooses the second, third, etc. best 5′ end extension primer (e.g. if the first one that Osprey suggested failed in the lab). The sequence file itself can be in Staden's ‘Strand coverage’ format or in plain FastA format (if an assembly engine other than gap4 was used). When in the sequence polishing (disambiguation and quality control) mode, regions with ambiguities and single-strand coverage are broken down into per-strand problem spot lists. The candidate primers are checked downstream to upstream with the expectation that early successful candidates may allow a single sequencing reaction to also resolve other problem locations slightly downstream with good quality sequence (Figure 2). Those downstream problems, assumed to be resolved by the sequencing reaction, are removed from the problem list. Single-strandedness problems on both strands are checked. Because the strand of sequencing used for disambiguation is not usually essential, the remaining ambiguities are resolved last. If a plain sequence file is provided instead of a Staden assembly file, only ambiguities can be resolved.
Figure 2.
Range of candidate sequencing primers for elucidation including position T1 (e.g. an assembly ambiguity to resolve), with user-specified parameters. Candidates are checked from 3′ to 5′ within the range to minimize the number of primers required. An early candidate could also resolve T2 in the same reaction. U = number of unreadable bases, L = primer length, R = expected sequence read length, M = minimum number of bases to elucidate after T1.
In the linking mode, assembly information can be marked NO 3PRIME or NO 3PRIME to exclude the ends of some contigs. For example, the sp6 or t7 ends of cosmids should not be included in the list of walking candidates, as the sequence generated would only contain vector. Walking mode also requires a minimum assembly overlap length parameter as an anchor to ensure that the resulting sequence read starts within the contig on which it was calculated. The candidate range for primers is determined by the following formulae, using the variable definitions from Figure 2, plus O = required assembly overlap bases (walking mode only):
Primer range end = T − U − O − L
Primer range start = range end − R + M
Without the M parameter, the best walking primer may result in only five bases beyond the already existing sequence! If no candidate meets the M requirement, the user can relax parameters such as secondary binding.
Microarrays
PCR-based methods involve aqueous medium duplex formation of two DNA strands similar to the application of sequencing primers. The priming of the template genomic sequence for gene PCR can be optimized for maximum efficiency and specificity, but there is no control over the binding properties of the PCR-product with cDNAs in the experiment. Genes with high DNA identity (e.g. paralogs), may bind well to each other's PCR-product even though their primed ends are unique, therefore redundancy filtration of the targets prior to primer design is suggested. This issue does not exist for spotted oligo probes because secondary binding of the oligo to the organism's whole transcript set is checked. However, both microarray types may suffer from binding problems due to secondary structures in the probe, which may hide the bases that are complementary to the target oligo.
PCR-based arrays
For PCR-based cDNA microarrays (23), Osprey expects a tab-delimited file with each row containing a gene label, and location of the 5′ and 3′ ends of genes in the input sequence. We have developed a Web interface for generating this file, based on the genome analysis from the Web-based MAGPIE annotation interface (24). This information can also be imported from GenBank records and other data sources compatible with the popular Readseq sequence reformatting utility embedded in Osprey. The gene priming sites undergo the same primer selection process as sequencing primers, but the target range formulae described above are simplified to identify the primers closest to the 5′ and 3′ ends of the coding sequence. The size of the gene product can also be given a maximum value (which we will refer to as G) to ensure that the length of the selected PCR amplification region is within the viable elongation range of the PCR (typically 1500 bases). In the case of long genes, the oligo selection process keeps the 3′ anchor, and chooses 5′ sites within G bases of that. The 3′ selection bias is based on the tendency of 3′ sequence to be over-represented in cDNAs when using 3′ mRNA Oligo(dT) priming in the probe amplification process for eukaryotic microarrays.
Arrays based on directly spotted oligonucleotides
Probe design for directly spotted oligo microarrays involves a different set of restrictions on the location of the oligo within the gene. For prokaryotic microarrays, we propose that in order to maximize detection of reverse transcription of sample mRNA, the bias for probe selection should be towards the 5′ end of the gene if candidate probe sites are equivalent by all other design parameters. For prokaryotes, typically a random hexamer nucleotide mixture is used to prime reverse transcription to cDNAs from the mRNA. This may generate a slight bias towards cDNAs with 5′ sequence. We assume that the random hexamers will prime reverse transcription (i.e. only create cDNA for mRNA sequence upstream of the priming site, towards the gene's 5′ end) at random locations on the mRNA. Hexamer analysis from S.solfataricus P2 shows no compositional bias change between 5′ and 3′ ends of genes, lending support to the assumption that the cDNA transcription starts would be random. An oligo matching the very 3′ end of the gene will have less cDNA to bind to than if it was more upstream, but the extent of the transcriptional bias will depend on many factors such as the PCR conditions and the length of the gene. Because the amount of this bias is uncertain, Osprey considers it only after all other, established constraints. For eukaryotic microarrays, a 3′ hybridization site bias is maintained, since a poly(T) is used to prime reverse transcription starting at the gene's 3′ mRNA poly(A) tail. Checks for secondary binding are restricted only to include transcribed sequences in the genome.
Other thermodynamic caveats exist for spotted oligo microarrays: Forman et al. (25) describe several discrepancies between standard models of aqueous thermodynamics and observed duplex formation for photolithographed oligos. Photolithographed probe fabrication is patented, and not within the scope of Osprey's usage, but all spotted oligos are likely subject to many of the same effects. No software for calculating of directly spotted oligos, such as PRIMEGENS (26), is documented to account for non-aqueous conditions. There is evidence for an approximately linear relationship between aqueous and fixed probe interaction thermodynamics, described by the authors of ProbeSelect (27). Formulae for fixed probes are not yet established, therefore we use the aqueous model as a guide.
Selection of optimal oligonucleotides
Osprey incorporates a series of fitness tests, in the following order: melting temperature, dimer potential, hairpin potential and secondary (non-specific) binding. Ordered from computationally simple to computationally expensive, the tests filter out unsuitable candidates as quickly as possible. Osprey distinguishes itself from existing programs in the way constraints are managed, computation is parallelized, problem sequences are dealt with, and secondary matches are found.
Thermodynamics for DNA binding and melting
Osprey uses dangling mismatch thermodynamics and SantaLucia's NN model. The number of base mismatches is not necessarily a reliable indicator of duplex stability, but the correlation between the free energy of the structure (ΔG, a measure of duplex stability) and the respective melting temperature is certain. This well-known correlation is attributable to the fact that both the melting temperature and the free energy are calculated using the same entropy and enthalpy values. Free energy is enthalpy minus the product of entropy and the reaction temperature (the classic Gibbs formula). The melting temperature formula used in Osprey was taken from (28),
![]()
where ΔH° is the sum of empirically derived enthalpy values for NNs in the oligo, ΔS° is the sum of empirically derived entropy values for NNs in the oligo, R is the molar gas constant and CT is the concentration of the oligo. We allow the users to set the minimum and the maximum melting temperatures to specify an acceptable range for their particular lab applications.
Undesirable annealing and parameter relaxation
In order to maximize the target binding efficiency, the binding of the oligo to itself must be minimized. Secondary binding against sequence other than the targeted site will also confound results by either mispriming in the case of primers, or promiscuously binding in the case of microarray spotted oligos.
Osprey's main program provides a mechanism to start the oligo design with tight constraints, and slowly loosen them if no appropriate oligos are found. Two variables that can be adjusted automatically are melting temperature and oligo length. A spotted oligo microarray design may start with a temperature range of 78 ± 5°C, and an oligo length range of 70 ± 5 bases. All possible oligos with 70 bases and melting at exactly 78°C will be found, and checked for fitness. Targets without any candidates passing the tests will be checked for 70mers with melting temperatures of exactly 77 or 79°C. This process continues until all 70mers with 73 or 83°C are checked, followed by the same checks for 71mers and 69mers and so forth. Osprey will print the best-yet passing candidate when the iteration is finished. When all iterations are finished, the best of the suboptimal oligos will be shown for target sequences that did not have any satisfactory oligo within the required length and temperature range.
If no optimal oligo length is given, Osprey determines one from the input sequences, based on the desired temperature. Two main uses of this are (i) adapting to genes with unusual base composition compared to the bulk of the query set and (ii) when the user is unsure of a statistically suitable oligo length for the query set. The adaptability is constrained in order to remain relevant for the lab application: user-specified bounds on the oligo length are still respected in this adaptable optimum mode. Average NN values for entropy and enthalpy are calculated by weighting each NN pair's energies by its relative frequency in the input. The energy averages are multiplied by L in the melting temperature formula previously discussed, and we solve for the only unknown, L.·L + 1 (since there is one more base than the number of neighbor pairs) is the average length of oligos with the specified melting temperature. This determination is recalculated after every candidate selection iteration. This speeds up the overall search: length values unlikely to have many candidates with the right melting temperature are skipped over until more statistically probable lengths are checked.
To facilitate the design of oligos for problematic sequences, the ‘rejects’ output from one run of Osprey can directly be used as the input to the next. Users can easily rerun the rejected targets with less stringent parameters (e.g. shorter oligo length, increased tolerance for secondary or hairpin binding) iteratively until the reject list is empty, or the remaining sequences are considered impossible to accurately target within the parameters of the experiment.
Dimer formation
The primer is compared to itself in all possible overlap lengths to check that two copies of the primer will not bind to each other to form a duplex. All possible matches, including those with small bulges, are checked. To be conservative, where the bases are not complementary, the ΔSj and ΔHj values are set to zero. The maximum value in kcal/mol is user-determined; an upper bound of −10 kcal/mol has proven effective for sequencing primers, −13 kcal/mol for cDNA microarrays. By contrast, dimer energy is typically above −25 kcal/mol for random 22mers with their complementary DNA.
Hairpin formation
The ends of a single primer may bind to each other, forming a hairpin structure. Osprey calculates all possible loop and stem lengths where complementation occurs, as well as interior loops and bulges, ignoring sterically impossible configurations such as one or two base hairpin loops (29). Once again, an upper limit of −10 kcal/mol free energy has proven effective in Osprey-generated sequencing primers, −13 kcal/mol for cDNA microarrays.
A program from the popular M-fold (30) package, quikfold, can be used to confirm the absence of significant secondary structure. Osprey is configured to use this check by default, similar to other oligo design programs including OligoArray (7).
Secondary binding
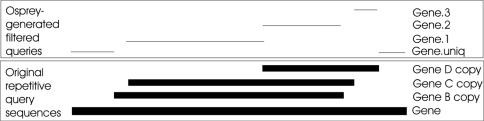
Formerly, Osprey employed an exhaustive linear software search of both the target and the secondary sequence with the thermodynamic criteria. This method was used for primers for the Sulfolobus sequencing project and the Candida microarray. While more accurate than BLAST-style pairwise identity searches, the method was slow for large data sets. Using the megablast program from the BLAST package, all repetitive elements larger than a user-defined threshold are now very quickly (<1 min for 3800 Sulfolobus genes on one CPU on a Sunfire 6800) identified in the query sequences. For whole genome analysis, the query file and the database are the same. If the user is iteratively searching for oligos using the ‘rejects’ from a previous run, the database remains the whole genome, while the query is just the sequences that do not yet have a suitable candidate. In either case, Osprey filters the query down to unique sequence, plus one copy of each repetitive section. This setup allows the secondary binding checks to be performed without interference from multi-copy elements. No user intervention or preprocessing of the data set is required, facilitating the use of Osprey with redundant data derived from GenBank and other sources. Figure 3 illustrates the repetitive sections' breakdown.
Figure 3.
Every segment of the gene that is either unique (noted by the .uniq suffix) or repeated by a distinct subset of the gene set is isolated for oligo design. To qualify for analysis, sections must be at least as long as the oligo length minus the maximum allowable repeat (default 20); hence some overlapping regions are not represented in the final oligos.
A novel computational method for the identification of secondary binding
Although the linear sequence search mode is still available in Osprey for users who cannot access accelerated hardware [both the TimeLogic™ Decypher® (http://www.timelogic.com) and Paracel™ GeneMatcher™ (http://www.paracel.com/) accelerators provide order-of-magnitude search time improvement for PSSMs], the current version of Osprey introduces a novel method of calculating and accelerating secondary binding checks using PSSMs. The models are compatible with the method established by Gribskov et al. (31). The NN modeling of thermodynamics requires a search method that scores a base-pairing according to the surrounding bases, to account for the local helix interaction effects. A single pairwise match or mismatch, when combined with information about the base immediately upstream, can have one of 2 × 42, or 32 energy states. Pairwise scoring as found in the Smith–Waterman algorithm (32) and its heuristic approximations only allows one of four scores for a match or mismatch at any particular position. An adenosine match gets the same score, regardless of the neighboring bases; therefore such scoring is unsuitable for encoding the many neighbor states required.
Osprey uses Gribskov profiles rather than Hidden Markov Models (33) (HMMs), because the log-odds scores for a position in the HMM must add up to one, whereas the profile search mechanism can work with arbitrary scores. Appropriately setting the position-specific scores allows the raw profile score to encode the significant caloric values of the binding. The properties we can encode are listed as follows.
A match score is the molar caloric free energy contribution of the matched base and its 5′ neighbor, and a portion of the unified model's length-dependent salt concentration penalty.
a mismatch score includes the free energy contribution of (a) the matched 5′ neighbor and mismatched base (b) the matched 5′ neighbor and mismatched base on the opposite strand (c) discount for the NN contribution in the next position.
The gap insertion penalty reflects the NN free energy penalty for single base bulges in a duplex.
The start of the sequence encodes the unified model's self-complementarity penalties if applicable. Mismatches in this position also encode mismatched end thermodynamics.
One extra state at each end of the profile sequence encodes dangling end thermodynamics in the case that the oligo matches to either terminus.
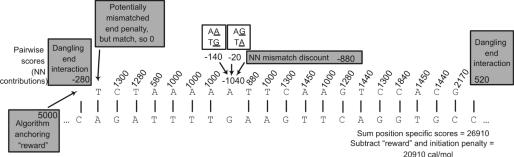
Figure 4 illustrates the scoring of a non-complementary duplex with all of the mentioned parameters. The per-position scores of the profile are added up as NN free energy contributions would be in traditional programs, except that the unified model's binding initiation penalty (∼1 kcal/mol) must be subtracted. The NN thermodynamics of single base bulges are taken from Ref. (34). A ‘reward’, equivalent to 5 kcal/mol, is given to the match of the 5′ end of the oligo (5 kcal/mol is larger than any NN mismatch score that would unanchor the 5′ match). This reward is required to ensure profile matches start at the 5′ end to properly include all the thermodynamic scores of the PSSM. The reward is then discounted from the final raw PSSM score to get the real thermodynamic score.
Figure 4.
Profile scoring used to encode caloric values from the NN thermodynamic model at 37°C with 1 M NaCl. The base and its 5′ NN determine the score for a match; C scores 1300 in positions 1, 10 and 17 of the top sequence (the oligo) because T preceeds it and it scores 1830 in position 18, because C proceeds it. The score position 8 is the sum of the two 5′–3′ mismatch NN values, minus the NN value overcounted in the following match. The AG/TA mismatch data is inverted to demonstrate that the actual 3′–5′ mismatch is equivalent to this standard 5′–3′ representation. The profile score summation, minus the standard initiation penalty of 1.0 kcal/mol is the duplex's free energy summation, 20.91 kcal/mol.
This representation reflects the thermodynamics of oligo duplexes, and compensates for dangling ends, as well as interspersed mismatches and bulges. Such a search is advantageous over a BLAST-type search because, unlike BLAST, the match, mismatch and gap scores are context sensitive (following the NN model). It also overcomes inherent limitations of the BLAST heuristic when dealing with short oligo sequences, such as missing DNA matches with gaps (duplex bulges), and interspersed mismatches. Due to these limitations, oligos where no apparent secondary binding was identified with BLAST may in fact show some using profiles (increased sensitivity). Also, candidates rejected due to a percentage similarity cutoff exceeded in BLAST may in fact not bind strongly to those sites when the NN thermodynamics are calculated (improved specificity). To provide a more intuitive measure of secondary binding to the user, the melting temperature of the best secondary match is calculated and displayed in the output.
Data parallelization
After repeats filtering and initial parameter setting, the calculation of a suitable microarray oligo for a gene is data-independent of the calculation for any other gene. This allows Osprey to simply split input gene lists into chunks that will be run in separate, concurrent processes. Surprisingly, none of the freely available software packages investigated while developing Osprey provide such parallelization explicitly. Computation is usually CPU-limited, therefore increasing the number of threads is beneficial up to the number of CPUs available. Melting temperature, dimer, hairpin and secondary binding checks are performed sequentially for each concurrent data chunk. When the Decypher® or GeneMatcher™ systems are used, the quikfold and PSSM searches are performed in parallel within the data chunk since the hardware PSSM searches put no CPU load on the system running Osprey.
RESULTS
A data set used to check free energy and melting temperature correlation in HyTher™ was also run against profiles to ensure that Osprey's new method results are in agreement with the best thermodynamic models. Free energy is measured with the profile rather than using melting temperature. Melting temperature calculation would require tracking two sets of scores, enthalpy and entropy, whereas profiles track one.
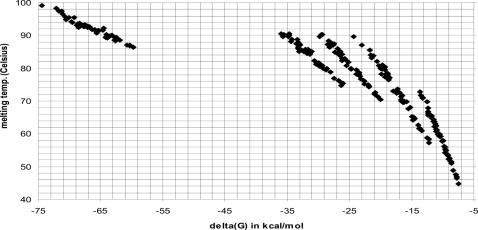
As expected, the results from the HyTher™ searches of matched and random oligos (Figure 5) indicate a strong correlation between melting temperature and ΔG, making ΔG a suitable candidate for selection and filtering of oligo specificity under experimental hybridization conditions.
Figure 5.
A dot plot of HyTher predicted free energy versus predicted melting temperature in 0.1 M NaCl for random exact matching oligos of lengths 10, 15, 20, 25 and 50 (forming clear groups from lower right to upper left).
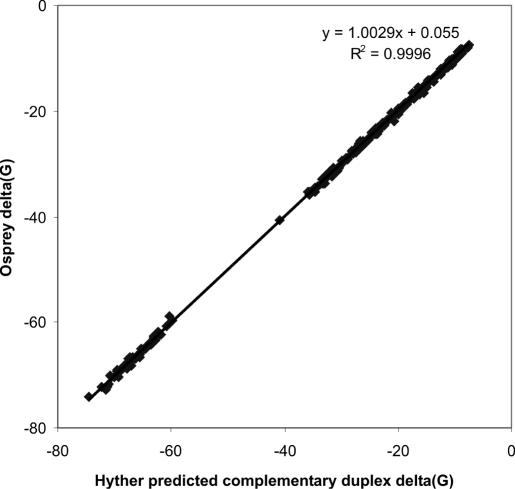
Results from the described profile constructs tested using the TimeLogic™ Decypher® system were found to strongly agree with HyTher™ predictions, as seen in Figure 6. This confirms that the new thermodynamics methods in Osprey reproduce best-of-breed results. The PSSM model is applied to secondary binding to improve sensitivity and selectivity in the sequence similarity search space.
Figure 6.
Prediction correlation between HyTher server (x) and Osprey (y) results for ΔG in kcal/mol using the 250 oligomers from Figure 5. R2 = 0.9996, with linear regression y = 1.0029x + 0.055, indicating Osprey's close conformity to the reference Unified Model predictions.
A spotted oligo probe array for S.solfataricus P2 was designed using the original linear search, then again using the PSSM searches with otherwise identical parameters, to compare the relative speed of the two approaches. The list of gene sequences was taken from the MAGPIE analysis for this genome. All three secondary search modes (namely Decypher®, GeneMatcher™ and internal linear search) were tested with otherwise identical search options. Decypher® and GeneMatcher™ modes take advantage of the parallel processing ability of these hardware accelerators by submitting not just a single candidate, but candidates for multiple sites of interest at once (Figure 1). The designated ideal oligo length was 70 bases, with a hybridization temperature target of 78 ± 5°C in 0.1 M NaCl for the 3775 (determined by MAGPIE) target genes. This temperature and salt concentration were chosen to match the other microarrays used in our facility, manufactured by Qiagen (http://oligos.qiagen.com/). Using the software linear search for secondary binding (58 h to complete) as a baseline of 1, speedup was approximately 8-fold for GeneMatcher2™, and 50-fold for Decypher®. Many factors can affect the potential speedup, including but not limited to the number of searches submitted concurrently to the hardware systems, and the specific hardware/software/firmware configuration. Our facility has a 4-board Decypher® system running on a SunSM v880, and a GeneMatcher2™ with 28000 ASIC CPUs, both maximal configurations.
The 500 primers (walking and polishing) designed for the S.solfataricus genome project using Osprey had a 15% failure rate. The presence of unknown repetitive sequences while walking on clones, and hidden errors in the sequence (compressions or low phred values), were a major cause of primer miscalculation. Tweaking the expected read length and maximum undesired free energy binding constraints minimized the number of primers required. Osprey is now also in use in the sequencing of the Aeromonas salmonicida genome, which has a 65% G + C content, as opposed to Sulfolobus's 35%. For clones with extension viability (i.e. any designed primers extend at all), Osprey primers have had an 89% success rate.
Through the use of the MAGPIE Web interface, the cDNA microarray for the human pathogen Candida albicans was designed (21), with initial successful expression profiles confirmed for 85% of the primer products spotted. We would expect this success rate to be even higher when using the updated thermodynamics, and a completed genome assembly. Sequence data was obtained from the Stanford Genome Technology Center (http://www-sequence.stanford.edu/group/candida).
DISCUSSION
Osprey attempts to give researchers a single program for a wide range of common oligo design tasks using all the standard exclusion filters. It also attempts to improve the rate of primer success with a novel method for specificity checking that is more sensitive and selective than pairwise alignment approaches. It automates the process of parameter relaxation as much as possible, and deals with repetitive sequence constructively.
Kampke et al. (35) suggest efficient primer design algorithms using dynamic programming. The authors concluded that ‘the full potential of mathematical calculation tools for this type of calculation has yet to be realized’. In this vein, we implemented the novel use of PSSMs to check for non-specific binding. PSSMs are a well-established mathematical tool in profiling biological sequences (33), and can be used to encode the thermodynamic profile of a sequence. Giving Osprey this option for checking secondary binding has clear advantages when one can harness the power of ultra-fast hardware-accelerated PSSM searches, such as those on the DeCypher® and GeneMatcher™ bioinformatics accelerators. As shown in the Sulfolobus microarray design, use of dedicated hardware components makes it practical, at least at the free energy level, to solve the ‘intractable problem’ (27) of simulating whole-genome thermodynamic interaction, rather than resorting to heuristics.
Looking forward from other groups' recent research and new Osprey methods, the use of dynamic computational methods clearly improves the efficiency of large-scale primer design processes. An approach which takes advantage of, rather than excludes, repeats (36) could be combined with the sensitivity and specificity of PSSMs to improve the multiple use of primers for the amplification of cDNAs. In a related application of sensitive secondary binding checks, Osprey could be directed to maximize secondary binding with a specific set of sequences, in order to create optimal oligos for microarrays to be used for multiple, related species. This could prove to be advantageous over the probabilistic methods introduced by OligoWiz (37).
New models of electrostatic effects on such microarrays (38) may lead to new thermodynamic parameters that improve accuracy based on linker molecule and substrate properties. If the NN parameters for such duplexes are well characterized in the future, Osprey's methods would be amenable to calculating these as well.
AVAILABILITY
Osprey is written as a set of C language code files, compiled into an executable program. The package does not rely on system-specific libraries, thus it should compile on most operating systems supporting C language compilers and Perl 5. The Web interface is a Perl wrapper around the command-line program. A Web version with hardware-accelerated searches is accessible at http://osprey.ucalgary.ca. Academic users can obtain the Osprey code base on an ‘as is’ basis by request to the corresponding author.
Acknowledgments
ACKNOWLEDGEMENTS
The Sun Center of Excellence for Visual Genomics is funded by ANPI, the Alberta Network for Proteomics Innovation; ASRA, the Alberta Science and Research Authority; WD, Western Economic Diversification; CFI, the Canadian Foundation for Innovation; Genome Canada; Genome Prairie; NSERC, the National Sciences and Engineering Council; Sun Microsystems Inc.; Fakespace Systems Inc.; and the University of Calgary. We would also like to acknowledge TimeLogic Inc.'s assistance in implementing DNA profile searches on the Decypher system.
REFERENCES
- 1.Wallace R.B., Shaffer,J., Murphy,R.F., Bonner,J., Hirose,T. and Itakura,K. (1979) Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: the effect of single base pair mismatch. Nucleic Acids Res., 6, 6353–6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomiuk S. and Hoffman,K. (2001) Microarray probe selection strategies. Brief. Bioinform., 2, 329–340. [DOI] [PubMed] [Google Scholar]
- 3.Chen B.Y., Janes,H.W. and Chen,S. (2002) Computer programs for PCR primer design and analysis. Methods Mol. Biol., 192, 19–29. [DOI] [PubMed] [Google Scholar]
- 4.Mrowka R., Schuchhardt,J. and Gille,C. (2002) Oligodb—interactive design of oligo DNA for transcription profiling of human genes. Bioinformatics, 18, 1686–1687. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen H.B. and Knudsen,S. (2002) Avoiding cross hybridization by choosing nonredundant targets on cDNA arrays. Bioinformatics, 18, 321–322. [DOI] [PubMed] [Google Scholar]
- 6.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rouillard J.-M., Herbert,C. and Zuker,M. (2002) OligoArray: genome-scale oligonucleotide design for microarrays. Bioinformatics, 18, 486–487. [DOI] [PubMed] [Google Scholar]
- 8.SantaLucia J. (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbour thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giegerich R., Meyer,F. and Schleiermacher,C. (1996) GeneFisher—software support for the detection of postulated genes. In States,D. et al. (eds), Proceedings of the Fourth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, pp. 68–77. [PubMed] [Google Scholar]
- 10.Breslauer K.J., Frank,R., Blocker,H. and Marky,L.A. (1986) Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA, 83, 3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rychlik W., Spencer,W.J. and Rhoads,R.E. (1990) Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res., 18, 6409–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rozen S. and Skaletsky,H.J. (2000) Primer3 on the WWW for general users and for biologist programmers. In Krawetz,S. and Misener,S. (eds), Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 13.Fotin A.V., Drobyshev,A.L., Proudnikov,D.Y., Perov,A.N. and Mirzabekov,A.D. (1998) Parallel thermodynamic analysis of duplexes on oligodeoxyribonucleotide microchips. Nucleic Acids Res., 26, 1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allawi H.T. and SantaLucia,J. (1997) Thermodynamics and NMR of internal G·T mismatches in DNA. Biochemistry, 36, 10581–10594. [DOI] [PubMed] [Google Scholar]
- 15.Allawi H.T. and SantaLucia,J. (1998) Thermodynamics of internal C·T mismatches in DNA. Nucleic Acids Res., 26, 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allawi H.T. and SantaLucia,J. (1998) Nearest neighbor thermodynamic parameters for internal G·A mismatches in DNA. Biochemistry, 37, 2170–2179. [DOI] [PubMed] [Google Scholar]
- 17.Allawi H.T. and SantaLucia,J. (1998) Nearest neighbor thermodynamics of internal A·C mismatches in DNA: sequence dependence and pH effects. Biochemistry, 37, 9435–9444. [DOI] [PubMed] [Google Scholar]
- 18.Peyret N., Seneviratne,P.A., Allawi,H.T. and SantaLucia,J. (1999) Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G, and T.T mismatches. Biochemistry, 38, 3468–3477. [DOI] [PubMed] [Google Scholar]
- 19.Bommarito S., Peyret,N. and SantaLucia,J. (2000) Thermodynamic parameters for DNA sequences with dangling ends. Nucleic Acids Res., 28, 1929–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.She Q., Singh,R.K., Confalonieri,F., Zivanovic,Y., Allard,G., Awayez,M.J., Chan-Weiher,C.C.-Y., Clausen,I.G., Curtis,B.A., De Moors,A. et al. (2001) The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl Acad. Sci. USA, 98, 7835–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nantel A., Dignard,D., Bachewich,C., Harcus,D., Marcil,A., Bouin,A.-P., Sensen,C.W., Hogues,H., van het Hoog,M., Gordon,P. et al. (2002) Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell, 13, 3452–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staden R. (1996) The staden sequence analysis package. Mol. Biotechnol., 5, 233–241. [DOI] [PubMed] [Google Scholar]
- 23.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 24.Gordon P., Gaasterland,T. and Sensen,C.W. (2001) Genomic data representation through images: MAGPIE as an example. In Sensen,C.W. (ed.), Genomics and Bioinformatics. Wiley-VCH, Weinheim, pp. 380–396. [Google Scholar]
- 25.Forman J.E., Walton,I.D., Stern,D., Rava,R.P. and Trulson,M.O. (1998) Thermodynamics of duplex formation and mismatch discrimination on photolithographically synthesized oligonucleotide arrays. ACS Symp. Ser., 682, 206–228. [Google Scholar]
- 26.Xu D., Li,G., Wu,L., Zhou,J. and Xu,Y. (2002) PRIMEGENS: robust and efficient design of gene-specific probes for microarray analysis. Bioinformatics, 18, 1432–1437. [DOI] [PubMed] [Google Scholar]
- 27.Li F. and Stormo,G.D. (2001) Selection of optimal DNA oligos for gene expression arrays. Bioinformatics, 17, 1067–1076. [DOI] [PubMed] [Google Scholar]
- 28.Borer P.N., Dengler,B., Tinoco,I.,Jr and Uhlenbeck,O.C. (1974) Stability of ribonucleic acid double-stranded helices. J. Mol. Biol., 86, 843–853. [DOI] [PubMed] [Google Scholar]
- 29.Freier S.M., Kierzek,R., Jaeger,J., Sugimoto,N., Caruthers,M.H., Neilson,T. and Turner,D.H. (1986) Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl Acad. Sci. USA, 83, 9373–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuker M. (2003) M-fold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res., 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gribskov M., McLachlan,A. and Eisenberg,D. (1987) Profile analysis: detection of distantly related proteins. Proc. Natl Acad. Sci. USA, 84, 4355–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith T.F. and Waterman,M.S. (1981) The identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 33.Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 34.LeBlanc D.A. and Morden,K.M. (1991) Thermodynamic characterization of deoxyribooligonucleotide duplexes containing bulges. Biochemistry, 30, 4042–4047. [DOI] [PubMed] [Google Scholar]
- 35.Kampke T., Kieninger,M, and Mecklenburg,M. (2001) Efficient primer design algorithms. Bioinformatics, 17, 214–225. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez R.J. and Skiena,S.S. (2002) Microarray synthesis through multiple-use PCR primer design. Bioinformatics, 18, S128–S135. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen H.B., Wernersson,R. and Knudsen,S. (2003) Design of oligonucleotides for microarrays and perspectives for design of multi-transcriptome arrays. Nucleic Acid Res., 31, 3491–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vainrub A. and Pettitt,B.M. (2003) Surface electrostatic effects in oligonucleotide microarrays: control and optimization of binding thermodynamics. Biopolymers, 68, 265–270. [DOI] [PubMed] [Google Scholar]