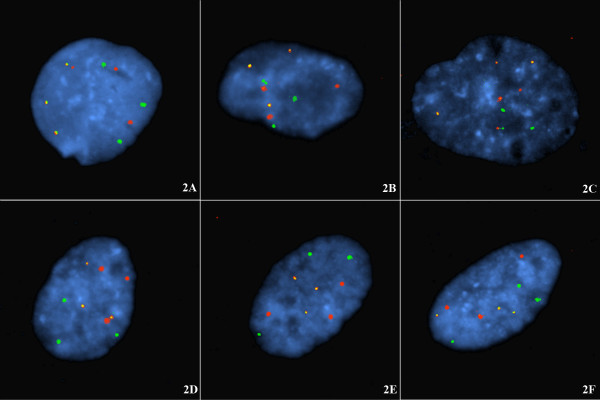
Figure 2.
A-F Chromosome homologues at G1 in nuclei of non-cycling cells are spatially arranged without respect to non-homologues. Same cell line and same cell harvest as the triploid cells probed in Fig 1. Two combinations of three subtelomeric probes (see Table 1 for clones) are shown hybridized to triploid cells. In fig 2A-C the nuclei are probed with three single subtelomere probes from 4p (spectrum orange); 18q (spectrum green) and 6p (both spectrum orange and spectrum green labels, i.e. yellow signal). In figs 2D-F, the nuclei are probed with three subtelomere probes labelled 5p (spectrum orange), 12q (spectrum green), and 20p (spectrum orange and spectrum green, i.e. yellow signal). Note: There was no segregation into haploids sets of chromosomes at G1 interphase. Homologues were regularly arranged without any defined relationship to non-homologous signal groups; i.e. haploid sets of interphase chromosomes distributed to separate nuclear regions do not appear to exist. Note also there is a low frequency of isolated non-homologous associations: between 4p and 6p (Fig 2A); between 6p and 18q (Fig 2C), and between 5p and 20p (Fig 2D).