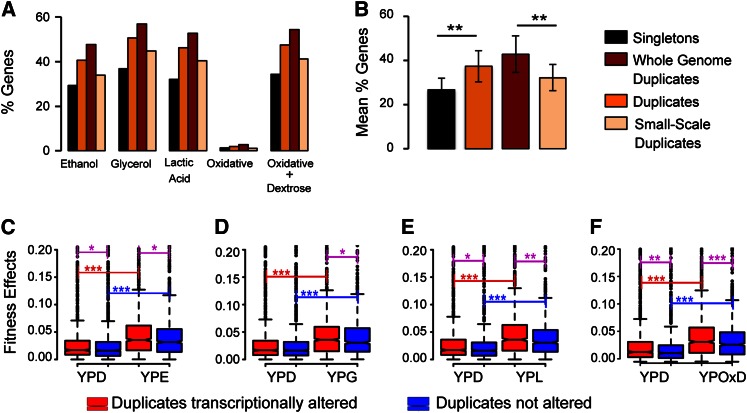
Figure 1.
Duplicated genes exhibit higher transcriptional plasticity than singleton genes and are involved in adaptation. (A) Percentage of genes with transcriptional flexibility when S. cerevisiae is grown under each of the five stress conditions tested in this study: ethanol stress, glycerol stress, acidic stress by lactate, oxidative stress, and oxidative stress in a medium supplemented with dextrose. (B) The mean percentage of genes of the categories’ singletons (black bar), duplicates (orange bar), duplicates generated by whole genome duplication (red bar), and duplicates generated by small-scale duplication (light yellow bar) with transcriptional alterations in the five stress conditions tested in this study. (C–F) We measured the contribution of transcriptionally altered duplicates to the fitness of S. cerevisiae under YPD and stress growth conditions using knock down gene data from Steinmetz et al. (2002). We then compared the fitness contribution of these altered duplicates (red boxes) with that of duplicates with no evidence for transcriptional plasticity under stress (blue boxes). These comparisons were performed for the sets of altered and not-altered duplicates identified under ethanol stress (C), glycerol stress (D), lactate stress (E), and oxidative stress supplemented with dextrose (F). Significant differences are indicated with *, **, and *** when the difference was significant at the levels of 0.01, 0.001, and 0.0001, respectively, using a Mann–Whitney U-test.