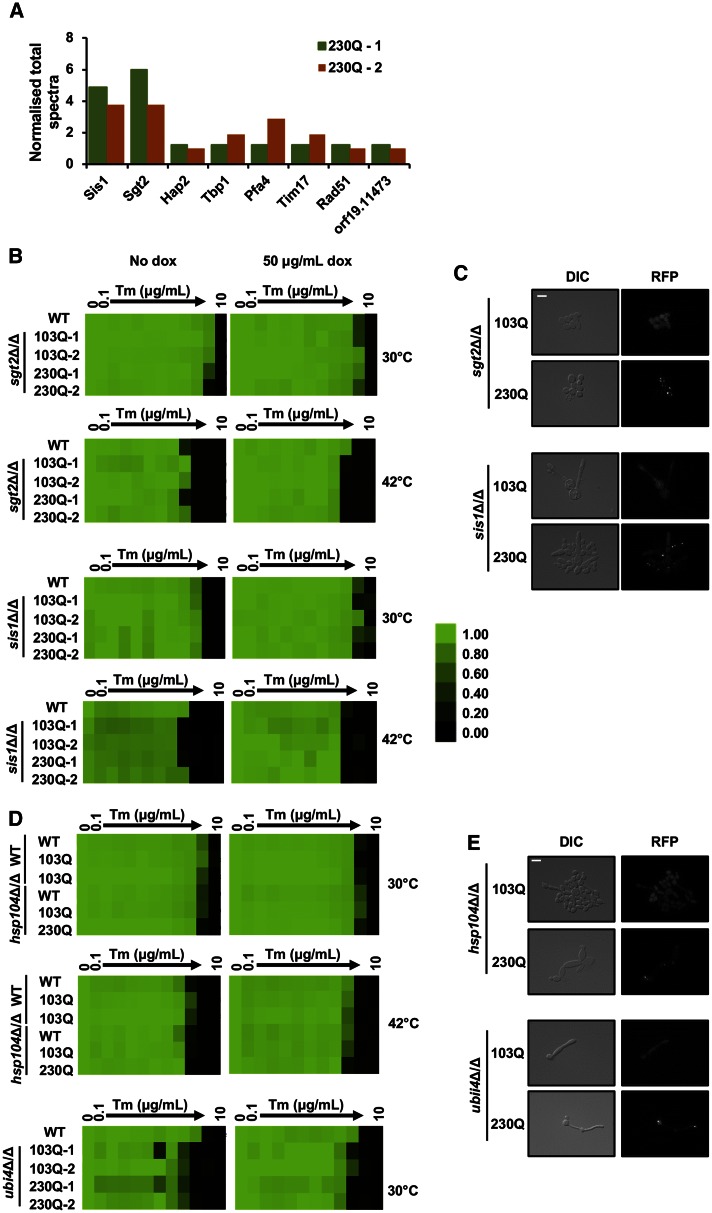
Figure 4.
Deletion of polyQ protein interactors does not increase toxicity or aggregation. (A) Protein interactors of 230Q. Histogram depicting the normalized total spectra count of proteins found to interact with 230Q in two biological replicates (230Q-1 and 230Q-2). (B) Toxicity in wild-type (WT) and independent sis1Δ/sis1Δ or sgt2Δ/sgt2Δ mutants, harboring 103Q or 230Q expression constructs, was assessed in the presence of a tunicamycin (Tm) gradient from 0 to 10 μg/ml, in twofold dilutions in YPD with or without 50 μg/ml doxycycline (dox) at 30 or 42°. Growth was measured after 48 hr of static incubation by absorbance at 600 nm and normalized relative to the no Tm control. For each strain, optical densities were averaged for duplicate measurements and displayed quantitatively using Treeview, as shown in the color bar. Data are representative for three biological replicates. (C) Live cell microscopy of 103Q and 230Q in sis1Δ/sis1Δ or sgt2Δ/sgt2Δ mutants grown at 42°. Scale bar, 10 μm. (D) Toxicity of WT and independent hsp104Δ/hsp104Δ or ubi4Δ/ubi4Δ mutants harboring 103Q or 230Q expression constructs was assessed in the presence of a Tm gradient from 0 to 10 μg/ml, in twofold dilutions in YPD with or without 50 μg/ml dox at 30 or 42°. Growth was measured after 48 hr of static incubation by absorbance at 600 nm and normalized relative to the no Tm control. For each strain, optical densities were averaged for duplicate measurements and displayed quantitatively using Treeview, as shown in the color bar. Data are representative for three biological replicates. (E) Live cell microscopy of 103Q and 230Q in hsp104Δ/hsp104Δ or ubi4Δ/ubi4Δ mutants grown at 42°. Scale bar, 10 μm. DIC, differential interference contrast; RFP, red fluorescent protein.