Figure 1.
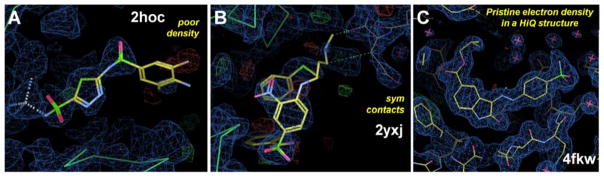
Crystal structures 2hoc and 2yxj have been used as benchmarking structures in other datasets. Their structures above show missing ligand density and influential symmetry contacts. Poor density gives unjustified ligand coordinates, and crystal-packing contacts force ligands and side chains to adopt incorrect orientations. (A) In 2hoc, the ligand has very poor density for roughly half the molecule, notably the ring on the right side of the figure. (B) In 2yxj, the tertiary amine of the ligand has poor density, and it is modeled in contact with the protein in the neighboring unit cell in the upper right. (C) Structure 4fkw was produced as part of CSAR’s experimental efforts, and it shows a ligand with pristine electron density for all atoms in the molecule. All side chains in the binding site are also well resolved, and even individual water molecules have good density. The ligand 62k (in the center of panel C) has an RSCC = 0.959. Ligands have yellow carbons; the protein backbone is a green chain; symmetry contacts have gray carbons. The electron density (2fo-fc map) is shown with standard blue contouring at 1.5σ. The errors (fo-fc map) are in the standard green and red colors.