Summary
Background
Improvements to prognostic models in metastatic castration-resistant prostate cancer have the potential to augment clinical trial design and guide treatment strategies. In partnership with Project Data Sphere, a not-for-profit initiative allowing data from cancer clinical trials to be shared broadly with researchers, we designed an open-data, crowdsourced, DREAM (Dialogue for Reverse Engineering Assessments and Methods) challenge to not only identify a better prognostic model for prediction of survival in patients with metastatic castration-resistant prostate cancer but also engage a community of international data scientists to study this disease.
Methods
Data from the comparator arms of four phase 3 clinical trials in first-line metastatic castration-resistant prostate cancer were obtained from Project Data Sphere, comprising 476 patients treated with docetaxel and prednisone from the ASCENT2 trial, 526 patients treated with docetaxel, prednisone, and placebo in the MAINSAIL trial, 598 patients treated with docetaxel, prednisone or prednisolone, and placebo in the VENICE trial, and 470 patients treated with docetaxel and placebo in the ENTHUSE 33 trial. Datasets consisting of more than 150 clinical variables were curated centrally, including demographics, laboratory values, medical history, lesion sites, and previous treatments. Data from ASCENT2, MAINSAIL, and VENICE were released publicly to be used as training data to predict the outcome of interest—namely, overall survival. Clinical data were also released for ENTHUSE 33, but data for outcome variables (overall survival and event status) were hidden from the challenge participants so that ENTHUSE 33 could be used for independent validation. Methods were evaluated using the integrated time-dependent area under the curve (iAUC). The reference model, based on eight clinical variables and a penalised Cox proportional-hazards model, was used to compare method performance. Further validation was done using data from a fifth trial—ENTHUSE M1—in which 266 patients with metastatic castration-resistant prostate cancer were treated with placebo alone.
Findings
50 independent methods were developed to predict overall survival and were evaluated through the DREAM challenge. The top performer was based on an ensemble of penalised Cox regression models (ePCR), which uniquely identified predictive interaction effects with immune biomarkers and markers of hepatic and renal function. Overall, ePCR outperformed all other methods (iAUC 0·791; Bayes factor >5) and surpassed the reference model (iAUC 0·743; Bayes factor >20). Both the ePCR model and reference models stratified patients in the ENTHUSE 33 trial into high-risk and low-risk groups with significantly different overall survival (ePCR: hazard ratio 3·32, 95% CI 2·39–4·62, p<0·0001; reference model: 2·56, 1·85–3·53, p<0·0001). The new model was validated further on the ENTHUSE M1 cohort with similarly high performance (iAUC 0·768). Meta-analysis across all methods confirmed previously identified predictive clinical variables and revealed aspartate aminotransferase as an important, albeit previously under-reported, prognostic biomarker.
Interpretation
Novel prognostic factors were delineated, and the assessment of 50 methods developed by independent international teams establishes a benchmark for development of methods in the future. The results of this effort show that data-sharing, when combined with a crowdsourced challenge, is a robust and powerful framework to develop new prognostic models in advanced prostate cancer.
Funding
Sanofi US Services, Project Data Sphere.
Introduction
Prostate cancer is the most common cancer among men in high-income countries and ranks third in terms of mortality after lung cancer and colorectal cancer.1 Of more than two million men diagnosed with prostate cancer in the USA over the past 10 years, roughly 10% presented with metastatic disease. For these men, the mainstay of treatment is androgen deprivation therapy, with a high proportion of response. However, responses are not durable, and nearly all tumours eventually progress to the lethal metastatic castration-resistant state. Although substantial improvements in outcome for men with metastatic castration-resistant prostate cancer have been achieved after approval of next-generation hormonal agents, an immunotherapeutic drug, a radiopharmaceutical agent, and a cytotoxic drug,2–10 how best to deploy these treatments has not been ascertained. Elucidation of variables associated with patients’ outcomes independent of treatment will facilitate the design of future trials by homogenising risk, thus enabling clinical trial questions to be answered more rapidly because smaller sample sizes will be needed.
Prognostic models in metastatic castration-resistant prostate cancer have been described11–13 using baseline variables from independent cohort studies. A 2014 prognostic model for metastatic castration-resistant prostate cancer14 included eight clinical factors predictive of overall survival: Eastern Cooperative Oncology Group (ECOG) performance status; disease site; use of opioid analgesics; lactate dehydrogenase; albumin; haemoglobin; prostate-specific antigen; and alkaline phosphatase. Can innovative models with improved performance be developed through a systematic search using data-driven approaches while providing insights into biological aspects of the disease that affect patients’ outcomes? An example of a novel clinical factor that is underexplored in contemporary prognostic model development is interaction effects between clinical variables, even though interactions between genetic variants are used widely and known to improve genetic-based risk prediction and patients’ stratification.15,16
Here, we present results from the prostate cancer DREAM (Dialogue for Reverse Engineering Assessments and Methods) challenge—an open-data, crowdsourced challenge in metastatic castration-resistant prostate cancer. A major contribution to this effort was removal of privacy and legal barriers associated with open access to phase 3 clinical trial data17 by Project Data Sphere—a not-for-profit initiative of the CEO Roundtable on Cancer’s Life Sciences Consortium that broadly shares oncology clinical trial data with researchers. The challenge was designed to accomplish two goals. First, we aimed to leverage open clinical trial data, enabling a community-based approach to identify the best-performing prognostic model in a rigorous and unbiased manner. Second, participating teams aimed to develop predictive models to both validate previously characterised predictive clinical variables and discover new prognostic features. Consistent with the mission of DREAM, all challenge data, results, and method descriptions from participating teams are available publicly through the open-access Synapse platform.
Methods
Trial selection
In April 2014, the DREAM challenge organising team reviewed all existing and incoming prostate cancer trial datasets (comparator arm only) in Project Data Sphere and selected four trials, which were the source of training and validation datasets for the DREAM challenge—ASCENT2,18 MAINSAIL,19 VENICE,20 and ENTHUSE 33.21 All four trials were randomised phase 3 clinical trials in which the comparator arm consisted of a docetaxel regimen and overall survival was the primary endpoint. These four trials also had similar inclusion and exclusion criteria: eligible patients were aged 18 years and older, had progressive metastatic castration-resistant prostate cancer, were chemotherapy-naive, and had an ECOG performance status of 0–2. Further details of inclusion and exclusion criteria for each trial are provided in the appendix (p 3). The patient-level trial datasets were deidentified by data providers and made available for the DREAM challenge through Project Data Sphere. No institutional review board approval was needed to access data.
Patient populations
We compiled training datasets from the comparator arms of ASCENT2, MAINSAIL, and VENICE. ASCENT218 is a randomised open-label study assessing DN-101 in combination with docetaxel. Patients with metastatic castration-resistant prostate cancer were randomly assigned either docetaxel and prednisone (comparator arm) or docetaxel and DN-101, stratified by geographical region and ECOG performance status. MAINSAIL19 is a randomised double-blind study to assess efficacy and safety of docetaxel and prednisone with or without lenalidomide in patients with metastatic castration-resistant prostate cancer. Participants were randomly assigned to either docetaxel, prednisone, and placebo (comparator arm) or lenalidomide, docetaxel, and prednisone. Stratification of patients in MAINSAIL was done based on ECOG performance status (0–1 vs 2), geographical region (USA and Canada vs Europe and Australia vs rest of world), and type of disease progression after hormonal treatment (rising prostate-specific antigen only vs tumour progression). VENICE20 is a randomised double-blind study comparing the efficacy and safety of aflibercept versus placebo, in which patients with metastatic castration-resistant prostate cancer were randomly assigned either docetaxel, prednisone or prednisolone, and placebo (comparator arm) or docetaxel, prednisone or prednisolone, and aflibercept. Participants were stratified by baseline ECOG performance status (0–1 vs 2). The validation dataset was from the ENTHUSE 33 trial,21 a double-blind study in which patients with metastatic castration-resistant prostate cancer were randomly allocated (1:1) either docetaxel and placebo (comparator arm) or docetaxel with zibotentan, stratified by centre.
Data curation
The original datasets from Project Data Sphere contained patient-level raw tables that conformed to either Study Data Tabulation Model (SDTM) standards or company-specific clinical database standards. To optimise use of these data for the DREAM challenge, we compiled the four sets of trial data into a set of five standardised raw event-level tables, meaning all four clinical trials were combined into the same tables based on laboratory values, medical history, lesion sites, previous treatments, and vital signs. Including patients’ demographic information, these tables presented most measurements made for the patient in that category. To summarise these data on a per-patient level, we created a core table, distilling the raw event-level tables and patients’ demographics into 129 clinically defined baseline and outcome variables. Full details of the data curation process are provided in the appendix (pp 3, 4).
We supplied participating teams with the full set of baseline and raw variables from the core and raw event-level tables. We encouraged challenge participants to derive additional baseline clinical variables from the five standardised raw event-level tables for modeling. We also provided teams with outcome variables for the ASCENT2, MAINSAIL, and VENICE trials, but we did not release the outcome variables for the ENTHUSE 33 trial because they would serve to independently evaluate the performance of models. The primary endpoint used for model development was overall survival, defined as the time from date of randomisation to the date of death from any cause.
We did principal component analysis to investigate systematic similarities or differences between the four clinical trials, using either all available variables or binary variables only. We visualised the principal component analysis by plotting the first principal component against the second principal component for all patients.
Further validation
After the DREAM challenge was completed using data from ENTHUSE 33 for method evaluation, we further validated the top-performing and reference models with data from a fifth trial, ENTHUSE M1,22 to assess whether the top-performing model could be used to stratify risk for patients with metastatic castration-resistant prostate cancer who received placebo alone and no docetaxel. ENTHUSE M1 is a randomised double-blind study to assess the efficacy and safety of 10 mg zibotentan in patients with metastatic castration-resistant prostate cancer (specifically, bone metastasis). By contrast with ENTHUSE 33, the ENTHUSE M1 trial included a comparator arm of placebo alone. Patients were randomly allocated (1:1) either zibotentan or placebo and were stratified by centre. The inclusion and exclusion criteria were similar to those used for ENTHUSE 33 except that patients in ENTHUSE M1 were pain free or mildly symptomatic. To be consistent for validation, curation of ENTHUSE M1 data followed the same process as was done for ASCENT2, MAINSAIL, VENICE, and ENTHUSE 33, resulting in a core table and five raw event-level tables.
Challenge procedures
The DREAM challenge was hosted and fully managed on Synapse, a cloud-based platform for collaborative scientific data analysis, through which all model predictions were submitted. The challenge was run in two phases (appendix pp 4, 17). First, teams were allowed to train and test their models in an open testing leaderboard phase. Second, teams were permitted one last submission to the final scoring phase, after which teams were scored and ranked. Accordingly, we split data from ENTHUSE 33 into two separate sets, consisting of 157 patients and 313 patients. The smaller dataset was used for the open testing phase and the larger dataset was used for the final scoring phase. Moreover, all reported performance values for the evaluated methods and all comparisons between the top-performing model and reference model used the larger set of data from the ENTHUSE 33 trial. The reference prognostic model for prediction of overall survival was a penalised Cox proportional-hazards model using the adaptive least absolute shrinkage and selection operator (LASSO) penalty.14
For method evaluation, we used the integrated AUC (iAUC)23 calculated from 6–30 months as our primary scoring metric. For robust determination of the best performing team or teams, we used Bayes factor analysis and randomisation test based on iAUC (appendix pp 4, 5). For each team, we calculated the Bayes factor to directly compare the performance of a model with the reference model; coefficients for the reference model were obtained from reported hazard ratios (HRs).14 Furthermore, we evaluated model predictions by plotting Kaplan-Meier curves, after dichotomising patients for each team separately by median risk score. We used the log-rank test to compare the two groups using the coxph function in the survival R package. We calculated CIs by inverting the Wald test statistic. The risk scores generated by each model have their own dynamic range; thus, we used the rankings of patients for scoring by iAUC or Kaplan-Meier analysis. Accordingly, we selected the median risk score as a means to compare different methods in a fair manner. A major goal of the challenge was to encourage teams to develop and test novel methods outside of standard survival analysis approaches; thus, risk score predictions across all teams varied in their range and distribution. A standard threshold could not be established fairly for all teams; therefore, we relied on rank-based scoring methods, including the iAUC, and stratifying risk scores based on the median. We also calculated other statistics, including median survival and 1-year and 2-year survival for the dichotomised high-risk and low-risk groups. We did hierarchical clustering on rank-normalised risk score predictions from all models in the challenge, using Euclidean distance and average linkage.
We used the ENTHUSE 33 dataset to assess the calibration of the top-performing model. We plotted the predicted survival probability based on the top-performing model against the observed survival proportions at 18, 24, 30, and 36 months. For each time cutoff, we divided the population into seven equally spaced categories based on the ranked predicted risk by the top-performing model. We then calculated the true survival proportion within each category and plotted it as a point estimate and 95% CI. A 45° line on the plots indicated perfect calibration.
The organisers of the DREAM challenge used SAS version 9.3 for data curation and R version 3.2.4 for statistical analyses. R packages used for challenge evaluation included survival version 2.38–3, ROCR version 1.0–7, timeROC version 0.3, and Bolstad2 version 1.0–28. The top-performing model also used glmnet version 2.0–5 and hamlet version 0.9.4-2.
Clinical trial data used in the prostate cancer DREAM challenge can be accessed online.24 Write-ups, model code, and predictions for all teams are reported in the appendix (pp 7, 8). Challenge documentation, including a detailed description of its design, overall results, scoring scripts, and the clinical trials data dictionary can be accessed via the Synapse platform.
Role of the funding source
Project Data Sphere had a collaborative role in design and logistics of the DREAM challenge but played no part in data collection, data analysis, and data interpretation or in the writing of this report. Sanofi US Services provided an in-kind contribution of human resources for curation of the raw datasets for the DREAM challenge and for clinical and scientific support of the challenge organisation, at the request of Project Data Sphere. Sanofi personnel participated in design of the DREAM challenge, in data analysis and data interpretation, and in writing of the report, but had no role in data collection. Raw clinical trial datasets for ASCENT2, MAINSAIL, and VENICE were available on the Project Data Sphere platform and were accessible by all registered users of Project Data Sphere, including all DREAM challenge participants and organisers, throughout the challenge. JG, TW, KKW, BMB, LS, KA, YX, FLZ, and JCC had access to raw data for ENTHUSE 33. JG, TW, KKW, LS, KA, FLZ, and JCC had access to raw data for ENTHUSE M1, during the post-challenge analysis. Data for ENTHUSE 33 and ENTHUSE M1 have been made freely accessible through the Project Data Sphere platform with publication of this report. The corresponding author had full access to all data in the study and had final responsibility for the decision to submit for publication.
Results
The overall DREAM challenge design is shown in figure 1, with full details in the appendix (p 4). The table presents baseline characteristics of patients in the five clinical trials included in this analysis. The training dataset included: 476 individuals from ASCENT2; 526 participants in MAINSAIL; and 598 men from VENICE. The validation dataset consisted of 470 patients from the ENTHUSE 33 trial; 528 men were initially enrolled to that trial but, because of regulatory restrictions in one country, data for 58 individuals were not made public through the challenge. The second validation dataset comprised 266 patients from ENTHUSE M1. Because of the same regulation restriction mentioned for ENTHUSE 33, some data were not provided to Project Data Sphere.
Figure 1. Study design.
Data were acquired from Project Data Sphere and curated centrally by the organising team to provide a harmonised dataset across the four studies. Three studies were provided as training data (ASCENT2, MAINSAIL, and VENICE) and the fourth (ENTHUSE 33) was the validation dataset. Teams submitted risk scores for ENTHUSE 33, then their predictions were scored and ranked using an integrated time-dependent area under the curve (AUC) metric.
Table.
Patients’ baseline characteristics
| ASCENT2 (n=476) | MAINSAIL (n=526) | VENICE (n=598) | ENTHUSE 33 (n=470) | ENTHUSE M1 (n=266) | |
|---|---|---|---|---|---|
| Age (years) | |||||
| 18–64 | 111 (23%) | 171 (33%) | 219 (37%) | 160 (34%) | 58 (22%) |
| 65–74 | 211 (44%) | 246 (47%) | 254 (42%) | 217 (46%) | 111 (42%) |
| ≥75 | 154 (32%) | 109 (21%) | 125 (21%) | 93 (20%) | 97 (36%) |
| ECOG performance score | |||||
| 0 | 220 (46%) | 257 (49%) | 280 (47%) | 247 (53%) | 196 (74%) |
| 1 | 234 (49%) | 247 (47%) | 291 (49%) | 223 (47%) | 70 (26%) |
| 2 | 22 (5%) | 20 (4%) | 27 (5%) | 0 (0%) | 0 (0%) |
| 3 | 0 (0%) | 1 (<1%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Missing | 0 (0%) | 1 (<1%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Metastasis | |||||
| Liver | 5 (1%) | 58 (11%) | 60 (10%) | 64 (14%) | 12 (5%) |
| Bone | 345 (72%) | 439 (83%) | 529 (88%) | 470 (100%) | 266 (100%) |
| Lungs | 8 (2%) | 74 (14%) | 88 (15%) | 56 (12%) | 13 (5%) |
| Lymph nodes | 163 (34%) | 298 (57%) | 323 (54%) | 208 (44%) | 80 (30%) |
| Analgesic use | |||||
| No | 338 (71%) | 347 (66%) | 419 (70%) | 339 (72%) | 256 (96%) |
| Yes | 138 (29%) | 179 (34%) | 179 (30%) | 131 (28%) | 10 (4%) |
| Lactate dehydrogenase (U/L) | 202 (176–250) | 210 (174–267) | NA | 213 (181–287) | 188 (170–219) |
| Missing | 13 (3%) | 1 (<1%) | 596 (100%) | 5 (1%) | 7 (3%) |
| Prostate-specific antigen (ng/mL) | 68·8 (24·2–188·4) | 84·9 (32·2–271·2) | 90·8 (30·8–260·6) | 99·6 (33·6–236·8) | 52·3 (17·3–153·0) |
| Missing | 1 (<1%) | 4 (1%) | 6 (1%) | 12 (3%) | 4 (2%) |
| Haemoglobin (g/dL) | 12·6 (11·6–13·6) | 12·7 (11·5–13·7) | 12·7 (11·7–13·5) | 12·5 (11·3–13·5) | 12·9 (12·2–13·7) |
| Missing | 3 (1%) | 10 (2%) | 0 (0%) | 4 (1%) | 2 (1%) |
| Albumin (g/L) | NA | 43 (41–45) | 42 (38–45) | 43 (40–46) | 43 (41–45) |
| Missing | 476 (100%) | 1 (<1%) | 16 (3%) | 2 (<1%) | 1 (<1%) |
| Alkaline phosphatase (U/L) | 113 (80–213) | 124 (81–265) | 135 (85–270) | 155 (98–328) | 130 (83–222) |
| Aspartate aminotransferase (U/L) | 24 (20–31) | 24 (19–31) | 25 (20–33) | 25 (20–33) | 24 (19–29) |
| Missing | 4 (1%) | 1 (<1%) | 8 (1%) | 3 (1%) | 3 (1%) |
Data are median (IQR) or number of patients (%). NA=not available. ECOG=Eastern Cooperative Oncology Group.
129 clinical baseline variables were measured for laboratory values, lesion site, previous medicines, medical history, and vital signs. When combined and assessed, the clinical variables for each trial were similar (appendix p 13), although when binary variables—mainly representing lesion sites—were judged separately, differences in clinical trials were recorded (appendix p 13). ASCENT2 had a lower frequency of patients with visceral metastases (1·1% liver and 1·7% lung) compared with individuals in the other three trials (10–14% liver, 11–15% lung). By contrast, the proportion of patients with bone metastases was high across the four trials (72–100%). Median follow-up differed among the four studies: 11·7 months (IQR 8·6–15·8) in ASCENT2; 9·2 months (6·4–13·1) in MAINSAIL; 21·1 months (12·9–29·6) in VENICE; and 15·3 months (10·9–20·8) in ENTHUSE 33. Risk profiles for each of the trials—specifically, mortality—were similar among the four trials (proportionality of hazards, p>0·5; appendix p 14). The proportion of patients who died in each trial was 138 (29%) of 476 in ASCENT2, 92 (17%) of 526 in MAINSAIL, 433 (72%) of 598 in VENICE, and 255 (54%) of 470 in ENTHUSE 33.
50 international teams—comprising 163 individuals—submitted predictions from their models to the challenge; with the reference model, the total number of models is 51. The distribution of all team scores by iAUC is shown in the appendix (p 15). The top-performing model was developed by a collaborative team from the Institute for Molecular Medicine Finland and the University of Turku. The method was based on an ensemble of penalised Cox regression (ePCR) models. The ePCR model extended beyond the LASSO-based reference model by using an elastic net to select additional correlated groups of clinical variables and their interactions, modelled as interaction terms (panel). The risk predictions from the trial-specific ensemble components were rank-averaged to produce the final ensemble risk score predictions and to avoid trial-specific variation.
The top-scoring ePCR model reported an iAUC of 0·791 and outscored all other teams, with a Bayes factor greater than 5, surpassing the threshold that defines significantly different performances (Bayes factor >3). The reference model achieved an iAUC of 0·743, with a significant difference in scores between the ePCR model and the reference model (Bayes factor >20). With a time-dependent AUC metric, the ePCR model outperformed the reference model at every timepoint, with the biggest difference in performance at later timepoints between 18 and 30 months (figure 2A). A median split of patients into low-risk and high-risk groups for the ePCR model resulted in a low-risk group comprising 156 patients and 56 deaths (median follow-up 27·6 months [IQR 18·2–31·9]) and a high-risk group containing 157 patients and 107 deaths (15·1 [8·5–20·1]). Similarly for the reference group, a low-risk group including 156 patients and 59 deaths (median follow-up 26·5 months [IQR 17·2–31·9]) and a high-risk group with 157 patients and 104 deaths (15·6 [8·6–21·8]) were generated. Kaplan-Meier analysis showed that low-risk and high-risk groups had significantly different overall survival in each model (ePCR, HR 3·32, 95% CI 2·39–4·62, p<0·0001; reference, 2·56, 1·85–3·53, p<0·0001; figure 2B, 2C). A full comparison is provided in the appendix (p 9). We assessed the calibration of the ePCR model by comparing predicted probabilities versus actual probabilities at multiple timepoints (appendix p 16).
Figure 2. Performance of ePCR model, using data from ENTHUSE 33.
(A) Time-dependent AUC was measured from 6 months to 30 months at 1-month intervals, reflecting the performance of predicting overall survival at different timepoints. (B, C) Overall survival was assessed by the Kaplan-Meier method, stratified by the median in the top-performing ePCR model (B) and the reference model (C). The log-rank test was used to compare risk groups. ePCR=ensemble of penalised Cox regression models. iAUC=integrated time-dependent area under the curve. HR=hazard ratio.
Figure 3 shows a network visualisation of the significant groups of variables identified in the ePCR model and their predictive relations, based on the importance of the model covariates and their interactions. Although many of the variables used in the reference model were also included in the ePCR model, aspartate aminotransferase was identified as a new important predictor. We also recorded a number of factors that were included as interaction terms, and of particular note were those reflecting the immunological or renal function of the patient. Prostate-specific antigen was an independent but weak prognostic factor that interacted strongly with lactate dehydrogenase and aspartate aminotransferase.
Figure 3. Projection of the most important variables and interactions in the ePCR model.
Automated data-driven network layout of the most significant model variables, according to their interconnections with other model variables. Node size and colour indicate the importance of the variable alone for prediction of overall survival and its coefficient sign, respectively. This importance was calculated as the area under the curve (AUC) of the penalised model predictors, as a function of penalisation parameter λ. Edge colour indicates the importance of an interaction between two model variables, with a darker colour corresponding to a stronger interaction effect. Coloured subnetwork modules annotate the variables based on expert curated categories. Variable and interaction statistics can be found in the appendix (pp 10, 11). ALB=albumin. ALP=alkaline phosphatase. AST=aspartate aminotransferase. BMI=body-mass index. ECOG=Eastern Cooperative Oncology Group. ePCR=ensemble of penalised Cox regression models. HB=haemoglobin. HCT=haematocrit. LDH=lactate dehydrogenase. PSA=prostate-specific antigen. RBC=red blood cell count.
In addition to identifying the top-performing model, the challenge also tested the other independent models, with 30 of 50 outperforming the reference model (Bayes factor >3; appendix p 15). We performed hierarchical clustering of risk scores from the 51 models to identify three distinct risk groups (figure 4A), with 98 patients (77 deaths) in group A (high risk), 131 patients (61 deaths) in group B (moderate risk), and 84 patients (25 deaths) in group C (low risk). Differences in overall survival among these three groups were significant (log-rank p<0·0001), with median overall survival of 12·9 months (95% CI 10·7–15·3) for group A, 20·8 months (18·3–25·6) for group B, and 27·7 months (26·6–not available) for group C (figure 4B).
Figure 4. Challenge meta-analysis.
(A) Hierarchical clustering of patients (Euclidean distance, average linkage) by rank-normalised prediction scores from all 51 models using the ENTHUSE 33 data. (B) Kaplan-Meier plot of survival probability for the three patient clusters from (A). Group A=high risk. Group B=moderate risk. Group C=low risk.
40 of 50 teams provided a list of common clinical factors that were incorporated into their final models; the frequencies with which a feature was reported as being important or significant in a team’s model are summarised in the appendix (p 18). The results not only confirmed the variables identified previously in the reference model but also highlighted several factors that were not. Of note, aspartate aminotransferase was included in more than half the team models. Other novel variables that were included in at least 15% of the models are total white blood cell count, absolute neutrophil count, red blood cell count, region of the world, body-mass index, and creatinine.
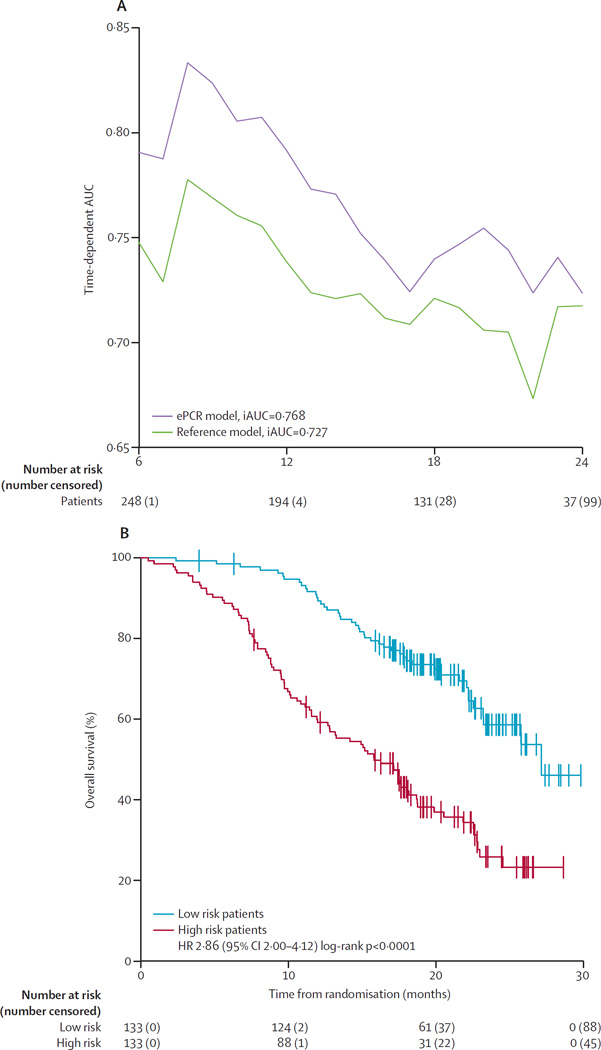
Application of the ePCR and reference models to the ENTHUSE M1 dataset showed model performances comparable with the primary challenge, with an iAUC of 0·768 for the ePCR model and 0·727 for the reference model (figure 5A). A median split of risk scores in the ePCR model led to a high-risk group of 133 patients, of which 45 were right-censored, and a low-risk group of 133 patients, of which 88 were right-censored. Kaplan-Meier analysis of the ENTHUSE M1 data showed significant separation of the high-risk and low-risk predicted patients (p<0·0001), with median survival of 15·8 months (95% CI 12·8–18·7) for high-risk patients and 27·1 months (23·2–not available) for low-risk patients (figure 5B).
Figure 5. Performance of ePCR model, using data from ENTHUSE M1.
(A) Time-dependent AUC was measured from 6 months to 24 months at 1-month intervals, reflecting the performance of predicting overall survival at different timepoints. The top-performing model (ePCR) is shown compared with the reference model. (B) Overall survival was assessed by the Kaplan-Meier method, stratified by median risk score. The log rank test was used to compare risk groups. ePCR=ensemble of penalised Cox regression models. iAUC=integrated time-dependent area under the curve. HR=hazard ratio.
Discussion
The prostate cancer DREAM challenge resulted in one prognostic model to predict overall survival significantly outperforming all other methods, including a reference model reported by Halabi and colleagues,14 and led to a network perspective of predictive biological variables and their interactions. The results from the top-performing team’s model pointed to important interaction effects with immune biomarkers and markers of hepatic function (potentially reflected in the increased amounts of aspartate aminotransferase) and renal function. The network visualisation of the prediction model suggests a complex relation and dependency structure among many of the predictive clinical variables. Many of these noted interactions, although not significant as independent variables, might be important modulators of key clinical traits—eg, haematology-related measurements such as haemoglobin and haematocrit. Although further investigation is necessary to determine the clinical implication of these associations and provide new insights into tumour–host interaction, these findings shed light on the complex and interwoven nature of prognostic factors on patients’ survival.
Open-data, crowdsourced, scientific challenges have been highly effective at drawing together large cross-disciplinary teams of experts to solve complex problems.25–30 To our knowledge, this DREAM challenge represented the first public collaborative competition31 to use open-access registration trial datasets in cancer with the intention of improving outcome predictions. In total, 163 individuals comprising 50 teams participated in the challenge, applying state-of-the-art machine learning and statistical modelling methods. The contribution of five clinical trial datasets from industry and academic institutions to Project Data Sphere, and their subsequent use in an open challenge, enabled the advancement of prognostic models in metastatic castration-resistant prostate cancer that up to now was not possible. Modellers had access to several independent clinical trial cohorts with subtle differences in eligibility that increased the diversity (heterogeneity) of the total patient population considered for model development. Access was also provided to data for 150 independent and standardised variables over the trials; by contrast, only 22 variables were considered by the reference model.14 The challenge resulted in creative data-mining approaches that used standardised raw event-level tables, which are rarely leveraged for prognostic model development, and enabled innovative clinical features to be derived for modelling. Several teams—including the top-performing team—made use of these event-level tables. Finally, evaluation of the 50 methods (validated by an independent and neutral party) provided the most comprehensive assessment of prognostic models in metastatic castration-resistant prostate cancer. These results are both a benchmark for future prognostic model development and a rich source of information that can be mined for additional insights into both patients’ stratification and the robustness of clinical predictive factors.
This study has shown the benefits of open data access at a time when clinicians, researchers, and the public are advocating for improved platforms and policies that encourage sharing of clinical trial data.32,33 Project Data Sphere has overcome major barriers to data sharing with support of data providers, to allow broad access to cancer clinical trial data. To researchers who are interested in leveraging open-access cancer trial data, this study represents a novel research approach that encompassed scientific rigor and a deep understanding of clinical data through effective collaboration of multidisciplinary teams of experts. The top-performing ePCR model was free of any a-priori clinical assumptions, with the exception of exclusion of non-relevant variables in early data curation. The data-driven modelling process identified automatically the best combination of predictors through cross-validation. Furthermore, the ePCR modelling process was fully agnostic to the variables used in the previous reference model; however, many of the same predictors were identified, in addition to novel ones. Such data-driven, unbiased modelling approaches can mine effectively the predictive variables and their combinations from large-scale and open clinical trial data.
The trials used here represent the standard of care at the time when the trials were done, which is a limitation of this study. Since 2010, several treatments have become available, for use both before and after first-line chemotherapy, and new trials have changed the way clinicians approach this disease.34 Abiraterone and enzalutamide—both approved for first-line treatment of metastatic castration-resistant prostate cancer—are not included within the scope of this challenge because of a limitation of control arm data; both COU-AA-3025 and PREVAIL10 have placebo or prednisone controls, and comparative trials using these agents as control have not been done. Accordingly, trial sponsors should be encouraged to contribute data from the experimental arm (particularly for approved drugs) to an active and engaged research community. Although sponsors are concerned that virtual comparisons might be made between treatments in experimental arms of different trials, there is far more benefit in leveraging these data to validate prognostic factors and models and to investigate intermediate clinical endpoints predictive of survival.
The DREAM challenge described here has shown that there is opportunity to further optimise prognostic models in metastatic castration-resistant prostate cancer using baseline clinical variables. For substantial advances beyond the work presented here, clinical trial data must be made available that reflects current advancements in treatment paradigms, including new data-capture techniques such as genomics, immunogenomics, and metabolomics that might more accurately describe the malignant state of the tumour and its microenvironment. Vital to either of these will be the need to share patient-level oncology data with the research community for the development of the next generation of prognostic and predictive models in cancer.
Supplementary Material
Research in context.
Evidence before this study
We searched PubMed between January, 2012, and July, 2015, with the terms “prognosis”, “overall survival”, “mCRPC”, and “docetaxel”. Our search yielded a 2014 study in which an updated prognostic model was described for metastatic castration-resistant prostate cancer that had been developed from the CALGB-90401 study (a randomised, double-blind, phase 3 clinical trial) and validated with data from the phase 3 ENTHUSE 33 trial. The study focused on a subset of clinical variables using datasets that were not in the public domain. Leveraging the wealth of data already generated from clinical trials is challenging on several fronts, but is complicated in particular by data access.
Added value of this study
Project Data Sphere is an independent not-for-profit initiative that aims to provide open access to historical patient-level data. The prostate cancer DREAM (Dialogue for Reverse Engineering Assessments and Methods) challenge is an open-data, crowdsourced competition to develop and assess prognostic models in metastatic castration-resistant prostate cancer. Using data from the comparator arms of four phase 3 clinical trials of chemotherapy-naive patients with metastatic castration-resistant prostate cancer, 50 independent teams—a diverse group of experts including biostatisticians, computer scientists, and clinical experts—developed prognostic models for the DREAM challenge, representing, to the best of our knowledge, the most comprehensive set of benchmarked models to date. The best-performing model was based on an ensemble of penalised Cox regression models that judged the prognostic value of interactions between predictor covariates and substantially outperformed the 2014 model. Strong support was provided for previously identified prognostic variables in the 50 models, and additional important variables were identified along with novel interactions between covariates. Data are available publicly through the Project Data Sphere initiative, and all method predictions and code are available for download through the Sage Bionetworks Synapse platform.
Implications of all the available evidence
Clinical trial data-sharing is both feasible and useful, and the DREAM challenge is an appropriate vehicle on which to build and rigorously assess prognostic or predictive models quickly, openly, and robustly. We established a new prognostic benchmark in metastatic castration-resistant prostate cancer, with applications in trial design and guidance for clinicians and patients. Robust and accurate prognostic predictors can be used to homogenise risk in clinical trials of metastatic castration-resistant prostate cancer and enable smaller trials for assessment of treatment effects.
Panel: Top-performing model construction in training datasets.
The top-performing model was based on an ensemble of penalised Cox regression models (ePCR), as shown in the equation. For each trial-specific ensemble component, the model estimation procedure identified an optimum penalisation parameter (λ), which controls for the number of non-zero coefficients in the prediction model, and simultaneously the regularisation parameter (α) with respect to the objective function:
Here, x are the predictors (clinical variables or their pairwise interactions), β are the model coefficients subjected to the absolute error and squared error penalisations (|β| and β2, respectively), p is the number of predictors, n is the number of observations, j(i) is the index of the observation event at time Ti, and Ri is the set of indices j for which yj≥Ti (patients at risk at time Ti), where yj is the observed death or right-censoring time. The set of indices Ri is redefined for each patient i using the above risk criterion incorporating y and T. With suitable regularisation, the penalised regression identifies an optimum balance between the model fit and top predictors, effectively generalising the Cox model for future predictions. To reduce the risk of overfitting and to avoid randomness bias in the binning, the final ensemble models were optimised using ten-fold cross-validation of the iAUC, averaged over multiple cross-validation runs. By modelling each trial individually as a separate ensemble component with different optima in the equation, we are able to account effectively for trial-specific variation (appendix p 12). The optimum parameters (penalisation λ and norm α) for each trial were first identified using cross-validation, after which the model coefficients (β) are estimated by optimising the above objective function.
Data processing entailed missing value imputation with a penalised Gaussian regression variant of the equation, with cross-validation when variables with non-missing values were used as predictors. Variables with missing values were inferred by training an optimum model with the non-missing variables and then imputing the missing values. Laboratory values were modelled as continuous variables. Data curation entailed unsupervised explorative analyses (appendix pp 5, 6, 12). ASCENT2 trial data were used in the imputation and unsupervised learning phases but were omitted from construction of the final supervised ensemble predictor, which was based on three components: MAINSAIL alone, VENICE alone, and their combination (appendix p 12). The final ensemble prediction was done by averaging over the ranks of the component-predicted risks for the ENTHUSE 33 dataset (appendix p 12). Averaging of risk score ranks was selected to be more robust to trial-specific variation and potential outliers. Full details of the model and its network visualisation are in the appendix (pp 5, 6, 12) with a list of chosen predictors (appendix pp 10, 11).
Acknowledgments
This report is based on research using information obtained from Project Data Sphere. Neither Project Data Sphere nor the owners of any information from the website have contributed to, approved, or are in any way responsible for the contents of this report. We thank the Sage Bionetworks Synapse team for the development and design of the DREAM challenge website. This work is supported in part by: the National Institutes of Health, National Library of Medicine (2T15-LM009451), National Cancer Institute (16X064; 5R01CA152301), Boettcher Foundation, Academy of Finland (grants 265966, 296516, 292611, 269862, 272437, 279163, 295504), Cancer Society of Finland, Drug Research Doctoral Programme (DRDP) at the University of Turku, and Finnish Cultural Foundation. Sanofi US Services provided an in-kind contribution of human resources for curation of raw datasets for the challenge and for clinical and scientific support of challenge organisation, at the request of Project Data Sphere.
CJS reports personal fees from Sanofi, Janssen, Astellas, and Bayer, outside the submitted work. FLZ and LS are employed by and have stock in Sanofi. KA is employed by and has stock in AstraZeneca, outside the submitted work. HIS reports non-financial support from AstraZeneca, Bristol-Myers Squibb, Ferring Pharmaceuticals, Medivation, and Takeda Millennium, outside the submitted work; consultancy fees from Astellas, BIND Pharmaceuticals, Blue Earth Diagnostics, Clovis Oncology, Elsevier (PracticeUpdate website), Genentech, Med IQ, Merck, Roche, Sanofi Aventis, WCG Oncology, and Asterias Biotherapeutics, outside the submitted work; and grants from Illumia, Innocrin Pharma, Janssen, and Medivation, outside the submitted work. GP reports grants from the Academy of Finland (decision number 265966), during the conduct of the study and outside the submitted work. SAK reports grants from the Academy of Finland (decision number 296516), during the conduct of the study and outside the submitted work. TDL reports research grants from the Finnish Cultural Foundation and Drug Research Doctoral Programme, during the conduct of the study; and a research contract from the National Cancer Institution, during the conduct of the study. TA reports grants from the Academy of Finland and the Cancer Society of Finland, during the conduct of the study; and a research contract from the National Cancer Institute, during the conduct of the study. OS reports grants and personal fees from Sanofi, outside the submitted work.
Footnotes
See Online for appendix
Contributors
JG, TW, JCB, ECN, TY, BMB, KA, TN, SF, GS, HS, CJS, CJR, HIS, OS, YX, FLZ, and JCC designed the DREAM Challenge. FLZ and LS led the efforts by Project Data Sphere to obtain and process the clinical trial data. TDL, SAK, GP, AA, TP, TM, and TA designed the top-performing method. JG, TW, TDL, KKW, JCB, ECN, GS, TA, FLZ, and JCC did the post-challenge data analysis and interpretation. HS, CJS, CJR, HIS, and OS assisted in clinical variable interpretation. All members of the Prostate Cancer Challenge DREAM Consortium (appendix pp 20–22) submitted prognostic models to the Challenge and provided method write-ups and the code to reproduce their predictions. JG, TW, TDL, HS, CJS, CJR, HIS, OS, TA, FLZ, and JCC wrote the report.
Declaration of interests
AA, JCB, BMB, JCC, SF, JG, TM, ECN, TN, TP, CJR, HS, GS, TW, KKW, TY, and YX declare no competing interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Tanimoto T, Hori A, Kami M. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 2010;363:1966. doi: 10.1056/NEJMc1009982. [DOI] [PubMed] [Google Scholar]
- 3.Berruti A, Pia A, Terzolo M. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;365:766. doi: 10.1056/NEJMc1107198. [DOI] [PubMed] [Google Scholar]
- 4.Fizazi K, Carducci M, Smith M, et al. Denosumab versus zoledronic acid for treatment of bone metastases in men with castration-resistant prostate cancer: a randomised, double-blind study. Lancet. 2011;377:813–822. doi: 10.1016/S0140-6736(10)62344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan CJ, Smith MR, de Bono JS, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368:138–148. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scher HI, Fizazi K, Saad F, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 7.de Bono JS, Oudard S, Ozguroglu M, et al. for the TROPIC investigators. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: a randomised open-label trial. Lancet. 2010;376:1147–1154. doi: 10.1016/S0140-6736(10)61389-X. [DOI] [PubMed] [Google Scholar]
- 8.Parker C, Nilsson S, Heinrich D, et al. Alpha emitter radium-223 and survival in metastatic prostate cancer. N Engl J Med. 2013;369:213–223. doi: 10.1056/NEJMoa1213755. [DOI] [PubMed] [Google Scholar]
- 9.Kantoff PW, Higano CS, Shore ND, et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 2010;363:411–422. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 10.Beer TM, Armstrong AJ, Rathkopf DE, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014;371:424–433. doi: 10.1056/NEJMoa1405095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halabi S, Small EJ, Kantoff PW, et al. Prognostic model for predicting survival in men with hormone-refractory metastatic prostate cancer. J Clin Oncol. 2003;21:1232–1237. doi: 10.1200/JCO.2003.06.100. [DOI] [PubMed] [Google Scholar]
- 12.Smaletz O, Scher HI, Small EJ, et al. Nomogram for overall survival of patients with progressive metastatic prostate cancer after castration. J Clin Oncol. 2002;20:3972–3982. doi: 10.1200/JCO.2002.11.021. [DOI] [PubMed] [Google Scholar]
- 13.Armstrong AJ, Garrett-Mayer ES, Yang Y-CO, de Wit R, Tannock IF, Eisenberger M. A contemporary prognostic nomogram for men with hormone-refractory metastatic prostate cancer: a TAX327 study analysis. Clin Cancer Res. 2007;13:6396–6403. doi: 10.1158/1078-0432.CCR-07-1036. [DOI] [PubMed] [Google Scholar]
- 14.Halabi S, Lin C-Y, Kelly WK, et al. Updated prognostic model for predicting overall survival in first-line chemotherapy for patients with metastatic castration-resistant prostate cancer. J Clin Oncol. 2014;32:671–677. doi: 10.1200/JCO.2013.52.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okser S, Pahikkala T, Airola A, Salakoski T, Ripatti S, Aittokallio T. Regularized machine learning in the genetic prediction of complex traits. PLoS Genet. 2014;10:e1004754. doi: 10.1371/journal.pgen.1004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Longo DL, Drazen JM. Data sharing. N Engl J Med. 2016;374:276–277. doi: 10.1056/NEJMe1516564. [DOI] [PubMed] [Google Scholar]
- 18.Scher HI, Jia X, Chi K, et al. Randomized, open-label phase III trial of docetaxel plus high-dose calcitriol versus docetaxel plus prednisone for patients with castration-resistant prostate cancer. J Clin Oncol. 2011;29:2191–2198. doi: 10.1200/JCO.2010.32.8815. [DOI] [PubMed] [Google Scholar]
- 19.Petrylak DP, Vogelzang NJ, Budnik N, et al. Docetaxel and prednisone with or without lenalidomide in chemotherapy-naive patients with metastatic castration-resistant prostate cancer (MAINSAIL): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet Oncol. 2015;16:417–425. doi: 10.1016/S1470-2045(15)70025-2. [DOI] [PubMed] [Google Scholar]
- 20.Tannock IF, Fizazi K, Ivanov S, et al. on behalf of the VENICE investigators. Aflibercept versus placebo in combination with docetaxel and prednisone for treatment of men with metastatic castration-resistant prostate cancer (VENICE): a phase 3, double-blind randomised trial. Lancet Oncol. 2013;14:760–768. doi: 10.1016/S1470-2045(13)70184-0. [DOI] [PubMed] [Google Scholar]
- 21.Fizazi K, Fizazi KS, Higano CS, et al. Phase III, randomized, placebo-controlled study of docetaxel in combination with zibotentan in patients with metastatic castration-resistant prostate cancer. J Clin Oncol. 2013;31:1740–1747. doi: 10.1200/JCO.2012.46.4149. [DOI] [PubMed] [Google Scholar]
- 22.Nelson JB, Fizazi K, Miller K, et al. Phase III study of the efficacy and safety of zibotentan (ZD4054) in patients with bone metastatic castration-resistant prostate cancer (CRPC) Proc Am Soc Clin Oncol. 2011;29 abstr 117. [Google Scholar]
- 23.Hung H, Chiang C-T. Estimation methods for time-dependent AUC models with survival data. Can J Stat. 2010;38:8–26. [Google Scholar]
- 24.Project Data Sphere. Prostate cancer DREAM challenge. [accessed Oct 21, 2016]; https://www.projectdatasphere.org/projectdatasphere/html/pcdc. [Google Scholar]
- 25.Costello JC, Stolovitzky G. Seeking the wisdom of crowds through challenge-based competitions in biomedical research. Clin Pharmacol Ther. 2013;93:396–398. doi: 10.1038/clpt.2013.36. [DOI] [PubMed] [Google Scholar]
- 26.Bender E. Challenges: crowdsourced solutions. Nature. 2016;533:S62–S64. doi: 10.1038/533S62a. [DOI] [PubMed] [Google Scholar]
- 27.Bansal M, Yang J, Karan C, et al. A community computational challenge to predict the activity of pairs of compounds. Nat Biotechnol. 2014;32:1213–1222. doi: 10.1038/nbt.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costello JC, Heiser LM, Georgii E, et al. A community effort to assess and improve drug sensitivity prediction algorithms. Nat Biotechnol. 2014;32:1202–1212. doi: 10.1038/nbt.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margolin AA, Bilal E, Huang E, et al. Systematic analysis of challenge-driven improvements in molecular prognostic models for breast cancer. Sci Transl Med. 2013;5:181re1. doi: 10.1126/scitranslmed.3006112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ewing AD, Houlahan KE, Hu Y, et al. Combining tumor genome simulation with crowdsourcing to benchmark somatic single-nucleotide-variant detection. Nat Methods. 2015;12:623–630. doi: 10.1038/nmeth.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saez-Rodriguez J, Costello JC, Friend SH, et al. Crowdsourcing biomedical research: leveraging communities as innovation engines. Nat Rev Genet. 2016;17:470–486. doi: 10.1038/nrg.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merson L, Gaye O, Guerin PJ. Avoiding data dumpsters: toward equitable and useful data sharing. N Engl J Med. 2016;374:2414–2415. doi: 10.1056/NEJMp1605148. [DOI] [PubMed] [Google Scholar]
- 33.Bierer BE, Li R, Barnes M, Sim I. A global, neutral platform for sharing trial data. N Engl J Med. 2016;374:2411–2413. doi: 10.1056/NEJMp1605348. [DOI] [PubMed] [Google Scholar]
- 34.Lewis B, Sartor O. The changing landscape of metastatic prostate cancer. Am J Hematol. 2015;11:11–20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.