Figure 3.
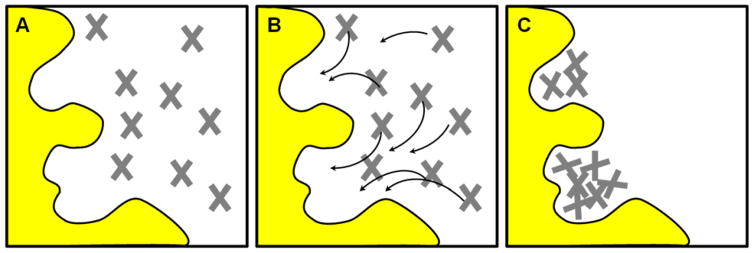
The MCSS method is based on energy minimization of probe molecules in the target binding site. Different probes map different interactions with the pocket, just like the GRID method, but the underlying mathematic implementation is different. (A) The binding site is initially flooded with thousands of probe molecules in random locations. (B) Each probe is treated independently, and its energy is based only on its interaction with the protein atoms. The arrows show the pathways toward the closest, local energy minima. The probes are systematically stepped “downhill” toward locations with more favorable energy. (C) No interactions between the probes are included in the calculations, so they can overlap one another at the end of the minimization. These clusters of probes map the favorable interaction sites on the protein surface. Further refinement of the probe locations and orientations uses a grid-based approach.