Abstract
Allosteric modulators, that exhibit no intrinsic agonist activity, offer the advantage of spatial and temporal fine-tuning of endogenous agonist activity, allowing the potential for increased selectivity, reduced adverse effects and improved clinical outcomes. Some allosteric ligands can differentially activate and/or modulate distinct signaling pathways arising from the same receptor, phenomena referred to as ’biased agonism’ and ’biased modulation’. Emerging evidence for CNS disorders with glutamatergic dysfunction suggests the metabotropic glutamate receptor subtype 5 (mGlu5) is a promising target. Current mGlu5 allosteric modulators have largely been classified based on modulation of intracellular calcium (iCa2+) responses to orthosteric agonists alone. We assessed eight mGlu5 allosteric modulators previously classified as mGlu5 PAMs or PAM-agonists representing four distinct chemotypes across multiple measures of receptor activity, to explore their potential for engendering biased agonism and/or modulation. Relative to the reference orthosteric agonist, DHPG, the eight allosteric ligands exhibited distinct biased agonism fingerprints for iCa2+ mobilization, IP1 accumulation and ERK1/2 phosphorylation in HEK293A cells stably expressing mGlu5 and in cortical neuron cultures. VU0424465, DPFE and VU0409551 displayed the most disparate biased signaling fingerprints in both HEK293A cells and cortical neurons that may account for the marked differences observed previously for these ligands in vivo. Select mGlu5 allosteric ligands also showed ‘probe dependence’ with respect to their cooperativity with different orthosteric agonists, as well as biased modulation for the magnitude of positive cooperativity observed. Unappreciated biased agonism and modulation may contribute to unanticipated effects (both therapeutic and adverse) when translating from recombinant systems to preclinical models.
Keywords: biased agonism, positive allosteric modulator, stimulus bias, metabotropic glutamate receptor 5
1. Introduction
In recent years, the metabotropic glutamate receptor subtype 5 (mGlu5), a G protein-coupled receptor (GPCR) ubiquitously expressed throughout the brain, has emerged as a promising drug target for multiple central nervous system (CNS) disorders, including schizophrenia, autism, Parkinson’s disease and depression (Gregory et al., 2013c; Niswender and Conn, 2010). Drug design efforts for mGlu5 are largely focused on targeting allosteric sites, that is, receptor binding sites topographically distinct from that of the endogenous ligand. Allosteric ligands offer the promise of increasing receptor selectivity, maximizing therapeutic effects and minimizing adverse effects. Further, modulation of orthosteric agonists by allosteric ligands without intrinsic agonism, allows for spatiotemporal fine-tuning of receptor responses – a favorable therapeutic mechanism within the complex CNS environment (Christopoulos and Kenakin, 2002; Leach et al., 2007; Melancon et al., 2012). Allosteric ligands that enhance receptor responses are called positive allosteric modulators (PAMs), while some allosteric compounds may also activate the receptor in the absence of endogenous ligand (PAM-agonists). Similar to orthosteric agonists, PAM-agonists do not have the benefit of temporal control of receptor activity. Allosteric ligands that diminish endogenous responses are termed negative allosteric modulators (NAMs), and those that neither increase nor decrease responses are referred to as neutral allosteric ligands (NALs) (Gentry et al., 2015). Discovery efforts for mGlu5 allosteric modulators have been particularly fruitful. Diverse modulator chemotypes recognize the ‘MPEP’ site, an allosteric pocket located within the receptor’s seven transmembrane bundle (Dore et al., 2014; Gregory et al., 2014; Gregory et al., 2013b; Wu et al., 2014). Of note, mGlu5 PAMs (classified based on glutamate stimulation of mGlu5-intracellular Ca2+ (iCa2+) mobilization in recombinant cells) have demonstrated efficacy in preclinical models of psychosis and cognition enhancement (Kinney et al., 2005; Rodriguez et al., 2010; Gregory et al., 2013a). However, recent studies have also reported adverse effects for select mGlu5 enhancers, such as seizure activity induced by VU0424465 (a PAM-agonist for mGlu5 mediated iCa2+ mobilization) (Rook et al., 2013) and neurotoxicity in rats treated with 5PAM523 (a pure PAM for mGlu5-iCa2+ mobilization stimulated by glutamate) (Parmentier-Batteur et al., 2014).
It has also become increasingly evident that, like many GPCRs, the mGlu5 receptor is pleiotropically coupled, activating multiple signaling pathways. In particular, while the receptor is predominantly coupled to Gq and subsequent changes in intracellular calcium (iCa2+) mobilization, mGlu5 couples to iCa2+-independent signaling pathways, such as extracellular signal-regulated kinases (ERK) 1 and 2 phosphorylation, cAMP, mTOR/PI3K, and can interact with, and modulate the activity of, other GPCRs and ion channels (Sengmany and Gregory, 2015). It is conceivable therefore that mGlu5 activation by chemically diverse ligands may engender unique receptor conformations, such that the receptor favors distinct subsets of physiological responses. This concept is known as ‘biased agonism’ (Kenakin and Christopoulos, 2013). Biased agonism offers the opportunity to design drugs that are tailored to activate the desired complement of receptor responses linked to therapeutic outcomes while avoiding those linked to adverse effects. Biased agonism is operative for orthosteric agonists at the related Group I mGlu1 receptor between G-protein and β-arrestin pathways in both recombinant and natively expressing cells (Emery et al., 2010; Emery et al., 2012; Hathaway et al., 2015). Several studies have suggested that mGlu5 biased agonism may be linked to distinct physiological responses (Gregory et al., 2013a; Gregory et al., 2012; Noetzel et al., 2013; Zhang et al., 2005). However, the vast majority of mGlu5 allosteric modulators have been classified as PAMs, PAM-agonists, NAMs or NALs, based on modulation of glutamate-mediated iCa2+ mobilization alone. To date, there have been no rigorous analyses of the mGlu5 signaling fingerprints of diverse allosteric modulators across different pathways. It is possible that preferential activation or inhibition of distinct mGlu5 signaling pathways could lead to different behavioral and/or toxicological outcomes.
One means to avoid the neurotoxicity and seizure liability associated with certain mGlu5 PAM and/or PAM-agonists (Rook et al., 2013; Parmentier-Batteur et al., 2014) and retain anti-psychotic and pro-cognitive effects may be via the development of biased mGlu5 modulators (Rook et al., 2015). Moreover, hitherto unappreciated bias may contribute to observations that markedly different doses of the same modulator are required for efficacy in different preclinical behavioral models. For example, DPFE, an mGlu5 PAM-agonist of glutamate-mediated iCa2+ mobilization and ERK1/2 phosphorylation in HEK293A cells, required vastly different doses to elicit cognition-enhancement versus reversal of amphetamine-induced hyperlocomotion in rats (Gregory et al., 2013a). Therefore, in the current study, we undertook the first rigorous analysis of mGlu5 bias for diverse PAMs in both HEK293A cells and primary cortical neuron cultures. Allosteric ligands were selected based on i) known interaction at the ‘MPEP’ allosteric site, ii) previous classification as PAMs or PAM-agonists in iCa2+ assays and iii) existing data in preclinical behavioral models. Propensity for biased allosteric agonism was determined relative to the orthosteric agonist DHPG across four signaling pathways that were amenable to assessment in both recombinant and neuronal cell cultures. We demonstrate that mGlu5 allosteric modulators display biased agonism and cooperativity, providing proof-of-concept that the development of biased mGlu5 allosteric modulators may be a means to tailor enhancement of mGlu5 activity to elicit therapeutically beneficial effects, such as anti-psychotic and cognition enhancement, while avoiding adverse effects.
2. Materials and Methods
2.1. Materials
Dulbecco’s modified Eagle’s medium (DMEM), Neurobasal and Opti- modified Eagle’s medium (Opti-MEM), Fluo-4-AM and antibiotics were purchased from Invitrogen (Carlsbad, CA). Fetal bovine serum (FBS) was purchased from Thermo Electron Corporation (Melbourne, Australia). IP-One HTRF® assay kit was purchased from Cisbio Assays, Genesearch (Arundel, QLD, Australia) and AlphaScreen detection beads were from PerkinElmer Life and Analytical Sciences. Select allosteric modulators N-cyclobutyl-5-((3-fluorophenyl)ethynyl)picolinamide (VU0403602), N-cyclobutyl-6-((3-fluorophenyl)ethynyl)picolin-amide (VU0360172), (R)-5-((3-fluorophenyl)ethynyl)-N-(3-hydroxy-3-methylbutan-2-yl)picolinamide (VU0424465), 1-(4-(2,4-difluorophenyl)piperazin-1-yl)-2-((4-fluorobenzyl)oxy)ethanone (DPFE), (5-((3-fluorophenyl)ethynyl)pyridin-2-yl)(3-hydroxyazetidin-1-yl)methanone (VU0405398) (4-fluorophenyl)(2-(phenoxymethyl)-6,7-dihydrooxazolo[5,4-c]pyridin-5(4H)-yl)methanone (VU0409551) were synthesized at Vanderbilt Centre for Neuroscience Drug Discovery as described previously (Gregory et al., 2013a; Gregory et al., 2012; Rook et al., 2013; Bridges et al., 2013; Rodriguez et al., 2010; Rook et al., 2015). N-(1,3-Diphenyl-1H-pyrazolo-5-yl)-4-nitrobenzamide (VU29), 3-Cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide (CDPPB) and 7-(Hydroxyimino)cyclopropa[b]chromen-1a-carboxylate ethyl ester (CPCCOEt) were purchased from Tocris Bioscience (Melbourne, Australia). Unless stated otherwise, all other reagents were purchased from Sigma-Aldrich (St. Louis, MO) and were of analytical grade.
2.2. Animals
All animal experiments and procedures were approved by the Monash Institute of Pharmaceutical Sciences Animal Ethics Committee (Protocol no. MIPS.2014.37). Asmu:Swiss outbred 8-week female wild-type mice were provided by the Monash Animal Research Platform (Clayton, Victoria, Australia). Time-mated females were humanely sacrificed and E16 embryos were recovered for primary cell culture.
2.3. Cell culture
HEK293A cells stably expressing wild-type rat mGlu5 at low levels (HEK293A-mGlu5-low), equivalent to those observed in primary cortical astrocytes (Noetzel et al., 2012) were maintained at 5% CO2, 37°C in DMEM supplemented with 5% FBS, 16mM HEPES and 500 μg/mL G418. The day before assays, cells were seeded onto poly-D-lysine coated, clear-bottom 96-well plates in assay medium (glutamine-free DMEM supplemented with 5% dialyzed FBS, 16 mM HEPES and 500 μg/mL G418) at a density of 40,000 cells/well, unless otherwise specified.
2.4. Primary cell culture
E16 Asmu:Swiss wild-type mice were decapitated, cortices dissected and neurons mechanically dissociated in Hank’s Balanced Salt Solution (HBSS; KCl 5.33 mM, KH2PO4 0.44 mM, NaHCO3 4.17 mM, NaCl 137.93 mM, Na2HPO4 0.34 mM, D-glucose 5.56 mM). Cortical neurons were then immediately plated on poly-D-lysine, FBS-coated clear-bottom 96-well plates in Neurobasal media, supplemented with 2 mM L-glutamine, 1 × B-27®, 50 U/mL penicillin, 50 μg/mL streptomycin, 1.25 μg/mL Fungizone® Antimycotic, at a density of 100 000 cells/well. Plates were maintained at 37°C and 5% CO2 for 5–7 days before experimentation, to allow growth of neurons.
2.5. Intracellular calcium mobilization
Intracellular calcium flux was measured as a change in fluorescence of the Ca2+ indicator dye, Fluo-4-AM, using a FlexStation I or III as described previously using a double-add paradigm, where allosteric ligands were added 1 min prior to orthosteric agonist (Gregory et al., 2012), with the exception of allosteric agonists, which were added simultaneously with orthosteric compound to avoid confounding effects of desensitization. Intracellular calcium mobilization was measured at room temperature (RT) for HEK293A-mGlu5-low cells and at 37°C for cortical neurons. For extracellular Ca2+-free experiments, CaCl2 was omitted from Ca2+ assay buffer (HBSS as above, with 2.5 mM probenecid, pH 7.4) and also supplemented with 1 mM EDTA. A 5-point smoothing function was applied to the raw fluorescence traces and peak fluorescence values (within 60 sec post-addition of ligand) normalized to the maximal response to either glutamate (HEK293A cells) or DHPG (cortical neurons).
2.6. Inositol monophosphate (IP1) accumulation assay
HEK293A-mGlu5-low or cortical neurons were washed with PBS and incubated for 1 h at 37°C with stimulation buffer (HBSS as above, with 20 mM HEPES, 30 mM LiCl2, 1.2 mM CaCl2, pH 7.4) before compound addition. After 1 h incubation, cells were lysed with Lysis Buffer (HTRF® IP-one assay kit) and IP1 levels determined using the HTRF® IP-one assay kit as per manufacturer’s instructions and fluorescence measured using the PHERAstar. Data were expressed as either fold over basal, or as a percentage of the maximal DHPG response.
2.7. ERK1/2 Phosphorylation
Receptor-mediated ERK1/2 phosphorylation was determined using an AlphaScreen-based ERK SureFire kit as described previously (Gregory et al., 2012). Cells were serum-starved using DMEM supplemented with 16 mM HEPES, for a minimum of 4 h for cortical neurons, and 6 h for recombinant cells. In HEK293A-mGlu5-low cells, the peak time for mGlu5-mediated ERK phosphorylation by all compounds was 5–7 min (Gregory et al., 2012; Rook et al., 2015b), while in cortical neurons, DPFE and VU0405398 peaked at 5 min, and all remaining modulators and DHPG peaked at 20 min (Supplementary Information). For interaction experiments in cortical neurons, allosteric modulators or vehicle were added 1 min prior to 20 min stimulation with DHPG. Assays were terminated by aspiration of ligand-containing medium and addition of 50 μL/well lysis buffer. Following 5 min shaking, 4 μL of lysate was transferred to a white 384 well ProxiPlate (PerkinElmer). Under low-light conditions, 7 μL AlphaScreen detection mixture (1:7 (v/v) activation buffer: reaction buffer; with 1:240 (v/v) acceptor and donor beads) was added to each well and incubated 1.5h at 37°C. AlphaScreen signal was measured using an Envision with standard AlphaScreen settings. Data were expressed as fold over basal levels of phosphorylated ERK.
2.8. Data analysis
Agonist-concentration response curves were fitted to a variable four-parameter logistic equation:
| (1) |
where bottom and top are lower and upper plateau levels of the concentration response curve respectively, n is the Hill coefficient, [A] is the molar concentration of agonist, and EC50 is the agonist concentration required to produce a half maximal response between top and bottom values (potency).
Allosteric modulation of glutamate or DHPG-mediated responses were fitted to the operational model of allosterism (Leach et al., 2007):
| (2) |
where [A] and [B] are the molar concentrations of orthosteric agonist, glutamate or DHPG, and allosteric modulator respectively. KA and KB are the equilibrium dissociation constants of the orthosteric agonist and allosteric modulator respectively, and α represents affinity cooperativity and β is a scaling factor that denotes the magnitude of effect an allosteric modulator has on the efficacy to an orthosteric agonist. Parameters τA and τB represent the respective ligand’s intrinsic ability to activate the receptor, while Em and n represent the maximal system response and the transducer slope respectively. KA values for orthosteric agonists were constrained to affinity estimates previously determined from inhibition binding assays (Gregory et al., 2012). Affinity cooperativity (α) was assumed to be neutral as validated previously (Gregory et al., 2012) and thus constrained to a value of 1, allowing estimates of β as a measure of cooperativity, that is the magnitude of effect a modulator has on orthosteric agonist potency and/or efficacy.
Biased agonism was quantified using an operational model of agonism (Kenakin et al., 2012):
| (3) |
where [A] is the agonist concentration, Em is the maximal response of the system, n is the transducer slope, and τ is the coupling efficiency of the agonist as defined by RT/KE, where RT is the receptor number and KE if the coupling efficiency of the system. From this equation the transduction coefficient log(τ/KA) a composite of both affinity and efficacy can be derived, which describes agonism and bias for a given pathway. A normalized transduction coefficient, Δlog(τ/KA), was consequently calculated using DHPG as the reference agonist. To compare different pathways (e.g. j1 vs j2), Δlog(τ/KA) values were evaluated between pathways by calculating the LogBias (Δ Δlog(τ/KA)):
| (4) |
Affinity, cooperativity and potency parameters were derived and represented as logarithmic mean ± SEM. Analysis of bias parameters was performed using one-way analysis of variance (ANOVA) with Tukey’s post-hoc test. Comparison of operational parameters between iCa2+ mobilization and IP1 accumulation in cortical neurons was determined using unpaired Student’s t-test.
3. Results
Eight allosteric ligands previously classified as either PAMs or PAM-agonists based on glutamate stimulation of mGlu5-mediated iCa2+ mobilization representing four distinct chemotypes were selected for this study to probe the potential for biased agonism and biased modulation of mGlu5 (Supplementary Fig. 1). In preclinical studies, the structurally related VU0403602 and VU0424465 have known adverse effect liability (Bridges et al., 2013; Rook et al., 2013) and provide a reference for modulators with undesirable properties. We also included two other ligands from this scaffold, VU0360172 and VU0405398, both of which have lower cooperativity with glutamate in iCa2+ assays, and minimal agonism compared to VU0403602 or VU0424465 (Gregory et al., 2012). CDPPB and VU29 were included as structurally distinct allosteric modulators from the same chemotype class, where CDPPB has been reported to be a PAM-agonist and VU29 a ‘pure’ PAM (Chen et al., 2007; Lindsley et al., 2004). CDPPB has previously been tested in vivo, showing efficacy in models of Huntington’s disease, fear extinction, psychosis and addiction (Cleva et al., 2011; Doria et al., 2015; Ganella et al., 2014; Gass et al., 2014; Uslaner et al., 2009). Recently, VU0409551 was described as a biased modulator on the basis that VU0409551 positively modulates mGlu5 activity in vitro but does not enhance mGlu5 modulation of NMDA current or NMDA receptor dependent plasticity in the hippocampus (Rook et al., 2015). Importantly, VU0409551 has efficacy in preclinical models of antipsychotic-like activity, cognition-enhancement and a genetic model of schizophrenia (Balu et al., 2016; Conde-Ceide et al., 2015; Rook et al., 2015). DPFE, a structurally distinct ligand relative to the three previous classes, was also included to offer further insight into the relationship between chemical scaffold and pharmacological outcome. As with CDPPB & VU0409551 behavioral studies have been conducted with DPFE (Gregory et al., 2013a; Peters et al., 2015), thereby allowing the potential to link in vivo efficacies with in vitro pharmacological fingerprints.
We first confirmed the pharmacological phenotypes for each of the eight allosteric modulators as potentiators of glutamate stimulation of mGlu5-mediated iCa2+ mobilization in HEK293A-mGlu5-low cells. In agreement with previous studies, compared to the orthosteric agonists glutamate and DHPG, VU0424465, VU0403602, and DPFE were efficacious partial allosteric agonists, achieving maximal responses 50–80% of the glutamate response. VU0360172, VU0409551 and CDPPB were also partial allosteric agonists. VU29 and VU0405398 had no appreciable allosteric agonist activity (Fig. 1A & B). The rank order of agonist efficacies was: glutamate = DHPG > VU0424465 > VU0403602 > DPFE ≥ VU0409551 > VU0360172 > CDPPB (see Supplementary Table 1 for Emax values). The rank order of agonist potencies (Supplementary table 1) was: VU0403602 > CDPPB ≥ VU0424465 > glutamate > VU0360172 > DHPG = VU0409551 > DPFE.
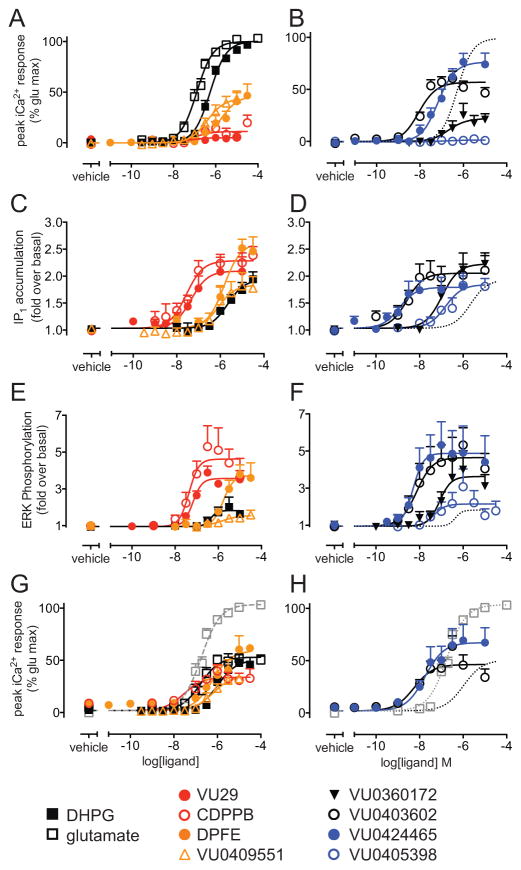
Figure 1. mGlu5 allosteric ligands have agonist activity for iCa2+ mobilization, IP1 accumulation and ERK phosphorylation in HEK293A-mGlu5-low.
Concentration-response curves for indicated mGlu5 orthosteric and allosteric ligands for iCa2+ mobilization (A & B), IP1 accumulation (C & D), ERK1/2 phosphorylation (E & F) and iCa2+ mobilization in the absence of 1.2mM CaCl2 (G & H; % glu max represents maximal glutamate response in the presence of 1.2mM CaCl2, curve from panel A shown in grey). In panels B, D, F and H, the DHPG curve is plotted as dotted lines for ease of reference. Responses were normalized to either the glutamate maximal response (iCa2+ mobilization) or represented as fold over basal (IP1 accumulation and ERK phosphorylation). IP1 and ERK1/2 phosphorylation experiments were performed in the presence of 10 U/mL GPT to minimize contribution of ambient glutamate. Data are expressed as mean + SEM of 3–17 experiments performed in duplicate, error bars not shown lie within the dimensions of the symbols.
All eight compounds had different degrees of positive cooperativity (logβ) with glutamate for iCa2+ mobilization, causing concentration-dependent increases in glutamate potency that approached a limit (Supplementary Figure 1). Modulator effects on glutamate concentration-response curves for iCa2+ mobilization were fitted to an operational model of allosterism to quantify the functional affinity (pKB) and efficacy modulation (logβ) for each of the ligands (Table 1). The resulting affinity and cooperativity estimates agree with those determined in previous studies (Bridges et al., 2013; Gregory et al., 2013a; Gregory et al., 2012; Manka et al., 2010).
Table 1.
Affinity and cooperativity estimates for allosteric modulation of orthosteric agonist-mediated iCa2+ mobilization and IP1 accumulation in HEK293A-mGlu5-low and cortical neurons. Data are mean ± SEM of 3–9 independent experiments performed in duplicate.
| HEK293A: glu-iCa2+ | HEK293A: DHPG-iCa2+ | Cortical: DHPG-iCa2+ | Cortical: DHPG-IP1 | |||||
|---|---|---|---|---|---|---|---|---|
| pKBa | logβb | pKB | logβ | pKB | logβ | pKB | logβ | |
| VU0424465 | 6.94±0.19 | 0.37±0.11 | 7.00±0.11 | 0.84±0.13c | n/a | n/a | n/a | n/a |
| VU0403602 | 7.62±0.17 | 0.97±0.15 | 7.23±0.34 | 1.06±0.12 | n/a | n/a | n/a | n/a |
| VU0360172 | 6.67±0.18 | 0.53±0.13 | 7.24±0.36 | 0.52±0.07 | 7.11±0.29 | 0.75±0.10 | 6.55±0.15 | 0.20±0.09d |
| VU29 | 6.87±0.22 | 0.13±0.09 | 6.67±0.28 | 0.81±0.08c | 5.70±0.45 | 1.07±0.24 | 6.51±0.19 | 0.29±0.04 |
| CDPPB | 6.58±0.25 | 0.69±0.16 | 6.87±0.17 | 0.72±0.08 | 6.61±0.25 | 0.66±0.15 | 6.81±0.41 | 0.25±0.12 |
| DPFE | 6.04±0.27 | 0.60±0.13 | 5.35±0.25 | 1.07±0.11c | 5.45±0.22 | 0.84±0.05 | 5.50±0.23 | 0.26±0.14d |
| VU0405398 | 7.00±0.12 | 0.31±0.06 | 6.97±0.24 | 0.57±0.06c | 5.37±0.29 | 0.56±0.08 | 6.46±0.13d | 0.11±0.03d |
| VU0409551 | 5.96±0.43 | 1.00±0.27 | 5.88±0.20 | 1.06±0.09 | 6.04±0.24 | 1.16±0.27 | 6.14±0.16 | 0.35±0.13d |
pKB, negative logarithm of the equilibrium dissociation constant determined using an operational model of allosterism.
logβ, logarithm of the efficacy modulation factor.
n/a denotes operational parameters not determined due to confounding agonism.
denotes p<0.05, comparing logβ values between glutamate and DHPG-stimulated iCa2+ mobilization.
denotes p<0.05, comparing respective operational parameters between iCa2+ mobilization and IP1 accumulation for cortical neurons, using an unpaired Student’s t-test.
3.1. Select mGlu5 PAMs show ‘probe dependence’ for potentiation of mGlu5-iCa2+ mobilization
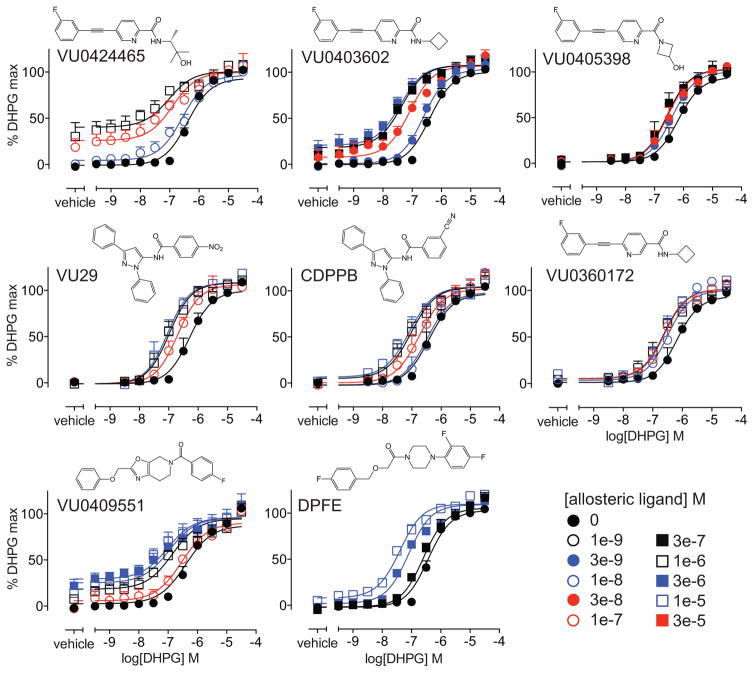
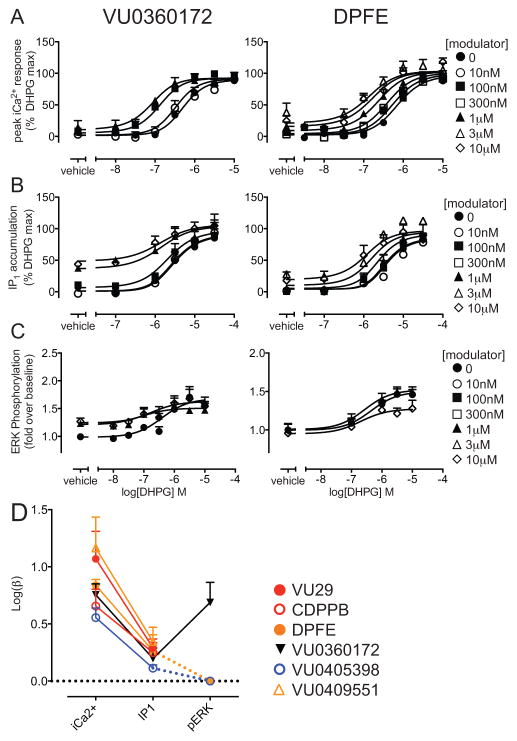
Although glutamate is the cognate agonist, it is a sub-optimal ligand to study the effects of mGlu5 alone in primary neuronal cultures due to the presence of multiple glutamate receptor subtypes and transporters in these native cells. Thus, we chose the more selective DHPG as the reference orthosteric agonist for assessing ligand pharmacology in primary neuronal cultures. We first confirmed that all eight allosteric ligands potentiated DHPG-mediated iCa2+ mobilization in HEK293A-mGlu5-low cells. DHPG concentration-response curves for iCa2+ mobilization in the absence or presence of different concentrations of each allosteric ligand (Fig. 2) were analyzed using the operational model of allosterism. As expected, affinity estimates were similar, irrespective of which orthosteric agonist was used (Table 1). Cooperativity estimates (logβ) for the interaction with DHPG were also similar to those determined with glutamate for VU0403602, VU0360172, VU0409551 and CDPPB. In contrast, the cooperativity between VU0424465, DPFE or VU0405398 with DHPG was ~2-fold higher than with glutamate. VU29 had ~5-fold higher positive cooperativity with DHPG compared to glutamate.
Figure 2. Allosteric modulation of DHPG-stimulated iCa2+ mobilization in HEK293A-mGlu5-low cells.
DHPG concentration-response curves for iCa2+ mobilization in the absence and presence of indicated concentrations of allosteric ligands. Interaction studies for DPFE, VU0409551, VU0424465, and VU0403602 were performed using simultaneous addition of both ligands, to minimize allosteric ligand-induced acute desensitization due to intrinsic agonist activity. VU29, VU0405398, CDPPB and VU0360172 were added 1 min prior to addition of glutamate. Data sets were globally fitted to an operational model of allosterism to estimate affinity and cooperativity. Curves represent the best fit of the data. Data are mean + SEM of n=3–10 experiments performed in duplicate. Error bars not shown lie within the dimensions of the symbol.
3.2. All allosteric ligands have direct agonist activity for mGlu5-mediated IP1 accumulation and ERK1/2 phosphorylation in HEK293A-mGlu5-low cells
We next assessed IP1 accumulation and ERK1/2 phosphorylation in response to the mGlu5 allosteric ligands, as alterations in these two pathways have been linked to diverse physiological outcomes (Bezprozvanny and Hayden, 2004; Ribeiro et al., 2010b; Tang et al., 2005); IP1 accumulation assay was assessed over a 1 h period. Glutamic pyruvic transaminase (GPT) 10 U/mL was added to reduce the effect of any ambient extracellular glutamate, which may confound observations of allosteric agonism (Supplementary Fig. 2). Inclusion of GPT reduced baseline levels of IP1 accumulation and phosphorylated ERK1/2 (pERK1/2) (Supplementary Fig. 2).
Relative to the reference orthosteric agonist, DHPG, all eight allosteric modulators were agonists for mGlu5-IP1 accumulation, achieving a similar maximal response to DHPG (Fig. 1C–D). All eight allosteric ligands stimulated pERK1/2, characterized by a transient peak (5–7min post-addition) that returned to baseline levels within 30min (Supplementary Fig. 2). Concentration-response curves for mGlu5-pERK1/2 constructed at the peak time point revealed VU0409551 and VU0405398 to be equally as efficacious agonists as DHPG (Fig. 1E–F). Consistent with a previous report (Gregory et al., 2012), VU29, CDPPB, DPFE, VU0360172, VU0424465 and VU0403602 elicited higher maximal responses for pERK1/2 than the orthosteric agonist. DHPG had significantly lower potency (7-fold) for IP1 accumulation relative to pERK1/2 and iCa2+ mobilization (Supplementary Table 1). Conversely, VU0424465 was 10–30 fold more potent in pERK1/2 and IP1 relative to iCa2+ assays, and VU0403602 had similar potencies for IP1 and iCa2+, but was ~5-fold more potent for pERK1/2. The potencies for VU0360172, CDPPB, VU0409551 and DPFE were similar across all three measures of receptor activation, as were VU29 and VU0405398 between pERK1/2 and IP1 accumulation. These differences in the rank orders of ligand potency for between different signaling pathways are suggestive of biased agonism.
3.3. mGlu5-mediated calcium mobilization is a composite of intracellular and extracellular calcium sources
Upon activation of Gq-coupled receptors, inositol phosphate hydrolysis is widely known to result in mobilization of intracellular calcium. Therefore it was unexpected that ligands that were agonists for IP1 had lower potency or efficacy in iCa2+ assays. However, it is also well-known that mGlu5 couples to various calcium ion channels (Sengmany and Gregory, 2015). Hence, iCa2+ mobilization detected upon receptor activation may be the combination of both extracellular calcium entering the cell via ion channels, and the calcium released from internal stores. To differentiate the sources of calcium, extracellular calcium was excluded from the buffer, thereby allowing measurement of calcium arising from intracellular stores. The maximal responses to both glutamate and DHPG were significantly reduced in the absence of 1.2 mM extracellular calcium, although potencies were unaffected (Fig. 1G). With the exception of CDPPB, which had increased efficacy in the absence of extracellular Ca2+, there was no significant change in the potency or maximal responses to DPFE, VU0409551, VU0403602 or VU0424465 between the differing calcium conditions with the mGlu5 allosteric ligands studied (Fig. 1G–H, Supplementary Table 1).
3.4. Assessment of agonist activity of mGlu5 allosteric ligands in cortical neurons
The same four measures of mGlu5 activity that we investigated in recombinant cells were next examined in cortical neurons to assess the potential of biased allosteric agonist activity in a native system (Fig. 3). Cortical neurons represent a physiologically relevant system to assess mGlu5 allosteric ligand activity, and can serve as a vital translational link for validating allosteric modulator pharmacology prior to preclinical assessments. To eliminate the influence of mGlu1, which is also expressed in the cortex, the mGlu1 selective negative allosteric modulator CPCCOEt 30 μM, was used in conjunction with DHPG for all experiments in cortical neurons. GPT had no effect on basal IP1 levels or allosteric agonism (Supplementary Fig. 3) and was not included in these experiments.
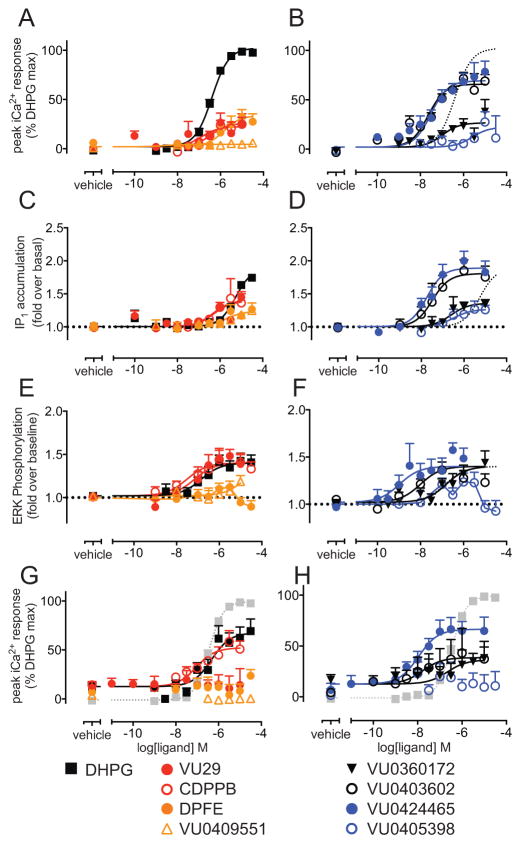
Figure 3. mGlu5 allosteric ligands have agonist activity for iCa2+ mobilization, IP1 accumulation and ERK phosphorylation in cultured cortical neurons.
Concentration-response curves for indicated mGlu5 orthosteric and allosteric ligands for iCa2+ mobilization (A&B), IP1 accumulation (C & D), ERK1/2 phosphorylation (E&F) and iCa2+ mobilization in the absence of 1.2mM CaCl2 (G & H; % DHPG max represents maximal DHPG response in the presence of 1.2mM CaCl2, curve from panel A is shown in grey). In panels B, D, F and H, the DHPG curve is plotted as a dotted line for ease of reference. Responses were normalized to either the glutamate maximal response (iCa2+ mobilization) or represented as fold over basal (IP1 accumulation and ERK phosphorylation). All assays were performed in the presence of 30 μM CPCCOEt to eliminate the contribution of mGlu1. Data are expressed as mean + SEM of 3–22 experiments performed in duplicate, error bars not shown lie within the dimensions of the symbols.
DHPG was ~10 fold less potent for IP1 accumulation relative to iCa2+ (in absence and presence of 1.2 mM extracellular Ca2+) and ERK1/2 phosphorylation (Supplementary Table 3). In iCa2+ mobilization assays relative to DHPG, VU0424465 and VU0403602 were nearly full agonists, whereas VU0360172, DPFE, VU29 and CDPPB were all partial agonists (Fig. 3A, B, Supplementary Table 2). VU0405398 also showed agonist activity, however, the maximal response was highly variable and restricted to concentrations above 3 μM. VU0409551 had no appreciable agonist activity for iCa2+ mobilization. The rank order of agonist efficacy: DHPG > VU0424465 = VU0403602 > CDPPB = VU29 = VU0360172 > VU0405398 ≫ VU0409551, was different to that observed in HEK293A cells.
For IP1 accumulation the relative efficacies of the allosteric ligands were similar to that observed for iCa2+ assays, with the exception of VU0409551 which had appreciable agonist activity for IP1 accumulation (Fig. 3C–D). The time course for mGlu5-mediated ERK1/2 phosphorylation by both orthosteric and allosteric ligands was characterized by sustained elevations or inhibition in cortical neurons, a markedly different profile to the transient response observed in HEK293A cells (Supplementary Figure 4). VU0424465, VU0403602, VU0360172, VU29, CDPPB and VU0409551 were all agonists for pERK1/2 in cortical neurons eliciting maximal responses to similar to DHPG (Fig. 3E–F). Allosteric agonism (iCa2+, pERK1/2 and IP1) in cortical neurons was inhibited by pre-exposure to the mGlu5 selective neutral allosteric ligand 5MPEP confirming that agonism was mediated via interaction with mGlu5 (Supplementary Fig. 5). Interestingly, while DPFE and VU0405398 were partial agonists for IP1 accumulation, they exhibited a bell-shaped concentration-response relationship for pERK1/2, elevating pERK1/2 at 1μM and dropping below basal at concentrations above 1μM (Fig. 3E–F). Similar to observations in HEK293A-mGlu5-low cells, the DHPG maximal response was significantly reduced in the absence of extracellular calcium (Fig. 3G). There were no significant differences in potency or maximal response of VU0424465, VU0403602, VU0360172, or CDPPB between differing calcium conditions (Fig. 3G–H). However, agonist activity by VU29, DPFE and VU0405398 was lost.
3.5. mGlu5 allosteric ligands are biased agonists
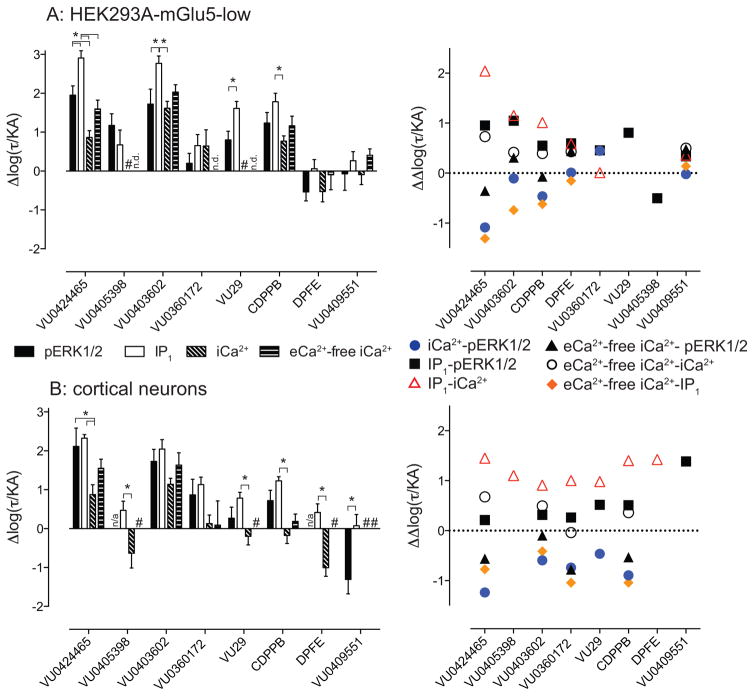
In order to rigorously and quantitatively assess the apparent bias of mGlu5 allosteric agonists in HEK293A-mGlu5-low cells and cortical neurons, observational bias between assays, and systems bias of the cell background should be minimized – thereby resulting in a more faithful estimate of true (conformationally-driven) biased agonism (Kenakin et al., 2012). From each assay, a transduction coefficient log(τ/KA) (Supplementary Table 4), that is, the composite of ligand efficacy and affinity, was derived as previously described (Kenakin et al., 2012) and normalized to the reference ligand DHPG (Δlog(τ/KA)). Subsequent normalization of Δlog(τ/KA) between pathways, ΔΔlog(τ/KA), allows for quantification of the degree of bias between pathways (Figure 4). Each allosteric ligand displayed a unique signaling bias fingerprint in HEK293A-mGlu5-low cells and neurons, although there were some notable trends. Relative to DHPG, VU0424465 showed significant bias away from iCa2+ mobilization and toward IP1 accumulation (110-fold) and ERK1/2 phosphorylation (9-fold) in HEK293A-mGlu5-low cells, a profile that was largely retained in neurons. VU0403602 was biased toward IP1 over pERK1/2 (11-fold) and iCa2+ mobilization (14-fold) in HEK293A-mGlu5-low cells, however this wasn’t reflected in the neurons. VU29 and CDPPB were biased (3–25 fold) toward IP1 over pERK1/2 and/or iCa2+ mobilization in both HEK293A cells and neurons. DPFE, VU0409551 and VU0405398 showed no discernible bias in HEK293A cells, but had distinct bias fingerprints in cortical neurons. VU0405398 and DPFE were biased toward IP1 over iCa2+ (12 and 26-fold respectively) coupled with biphasic pERK1/2 responses that could not be analyzed using the operational model. VU0409551 was the only ligand to show significant bias (24-fold) between IP1 and pERK1/2 in cortical neurons.
Figure 4. mGlu5 allosteric ligands are biased agonists relative to DHPG in HEK293A-mGlu5-low and cortical neurons.
The transduction coefficient (log(τ/KA)) was derived by applying equation 3 to agonist concentration-response curves in HEK293A-mGlu5-low (A, curves in Figure 1) and cortical neurons (B, curves in Figure 2) and normalized to DHPG (Δlog(τ/KA)). To calculate the degree of bias evident for different ligands between different pathways, Δlog(τ/KA) values were subtracted from one another ΔΔ ( log(τ/KA)) to determine Log bias factors. Data for Δlog(τ/KA) represent the mean ± SEM, whereas Log bias estimates are mean only. * denotes significantly different comparisons, p<0.05, one-way ANOVA with Tukey’s post-test.
3.6. CDPPB, DPFE, VU0405398 are biased allosteric modulators of DHPG in cortical neurons
We were unable to perform interaction studies between the modulators with DHPG to quantify affinity and cooperativity in HEK293A-mGlu5-low cells, because of the robust agonism for IP1 accumulation and ERK1/2 phosphorylation displayed by each allosteric ligand in their own right. However, in cortical neurons, a number of allosteric ligands had minimal or partial agonist effects, such that a sufficient response window remained to allow for allosteric interaction experiments with DHPG in this physiologically relevant system. Thus, the resulting DHPG concentration-response curves for IP1 and iCa2+ in the absence and presence of increasing concentrations of VU0360172, VU29, CDPPB, DPFE, VU0409551 or VU0405398 were constructed and analyzed using an operational model of allosterism (Fig. 5, Supplementary Fig. 6). Affinity estimates were similar for each allosteric ligand across the two pathways, with the exception of VU0405398, which had higher affinity in the IP1 assay (Table 1). For all six allosteric modulators, the magnitude of logβ was lower for IP1 versus iCa2+ (Fig. 5D); this reached significance for DPFE (3.8-fold), VU0360172 (3.6-fold), VU0409551 (6.5-fold) and VU0405398 (2.8-fold). Given that VU0360172, VU0405398 and DPFE were also partial agonists for ERK1/2 phosphorylation, we assessed modulation of the DHPG concentration-response curve by these three modulators. In cortical neurons, VU0360172 was a PAM-agonist for DHPG-stimulated ERK1/2 phosphorylation with similar affinity and cooperativity estimates (pKB: 6.49±0.20 and logβ: 0.69±0.18) to those determined in iCa2+ assays (Fig. 5D). In contrast, DPFE and VU0405398 showed a unique bell-shaped modulatory response in ERK1/2 phosphorylation. While 1 μM of each ligand potentiated DHPG responses, 10 μM of each ligand resulted in decreased DHPG maximal responses for ERK1/2 phosphorylation (Fig. 5).
Figure 5. mGlu5 allosteric ligands are biased modulators of DHPG responses in native cortical neurons.
DHPG concentration response curves for iCa2+ mobilization (A), IP1 accumulation (B) and ERK1/2 phosphorylation (C) in the absence and presence of VU0360172 and DPFE. Both VU0360172 and DPFE produced leftward shifts in DHPG potency for iCa2+ and IP1 assays. Data sets were globally fitted to an operational model of allosterism to estimate affinity and cooperativity. Curves represent the best fit of the data. In ERK1/2 phosphorylation assays, DPFE produced a leftward shift with 1 μM however reduced the maximal response of DHPG at 10 μM. D) Cooperativity estimates (Logβ) of allosteric ligands were plotted to enable comparison of modulation across pathways. For DPFE and VU0405398, which showed mixed modulatory activity, Log β values were plotted as zero to highlight the implicit bias; the absolute numerical value in this instance has no meaning. Data are mean ± SEM of n=3–9 experiments performed in duplicate. Error bars not shown lie within the dimensions of the symbol.
4. Discussion
Allosteric modulators of mGlu5 offer considerable therapeutic potential for the treatment of numerous psychiatric and neurological disorders. In particular, mGlu5 PAMs have demonstrated efficacy in preclinical models of psychosis, cognition-enhancement and NMDA receptor hypofunction (Ayala et al., 2009; Gregory et al., 2013a; Horio et al., 2013; Rook et al., 2015; Uslaner et al., 2009). However, a number of recent studies have reported adverse effects with select mGlu5 allosteric modulators (Rook et al.,, 2013; Parmentier-Batteur et al.,, 2014). Therefore in order to realize the therapeutic potential of mGlu5 allosteric modulators, there is a need to better understand the full scope of allosteric drug action. We tested the hypothesis that structurally diverse PAMs had unappreciated biased pharmacology. We found that mGlu5 allosteric ligands had diverse biased signaling fingerprints in both recombinant and native systems. Furthermore, this bias extended to modulation of orthosteric agonist activity, where the magnitude of cooperativity between allosteric and orthosteric ligands differed depending upon the measure of receptor activation. The prevalence of biased agonism and modulation for diverse mGlu5 PAM chemotypes highlights the paucity in our understanding of the functional consequences of mGlu5 allosteric modulation, and raises new avenues for therapeutic discovery. Armed with a better appreciation of the full scope of mGlu5 allosteric modulator action, it may be possible to link activation (and/or enhancement) of specific pathways to different physiological outcomes to promote therapeutic effects and avoid adverse effects.
In particular, many of the studied mGlu5 allosteric ligands, which previously were largely classified based on modulation of glutamate-stimulated iCa2+ mobilization, showed significant differences in coupling efficiency to iCa2+ mobilization, IP1 accumulation and ERK1/2 phosphorylation. Most strikingly, each allosteric ligand tested had greater efficacy for IP1 accumulation relative to iCa2+ mobilization. Therefore the ligands tested cannot be considered as pure mGlu5 PAMs, and do not posses the safety advantages of pure allosteric modulators, i.e., the advantage of spatiotemporal fine-tuning of receptor responses and saturable effects. Classically, iCa2+ mobilization is considered to be downstream of inositol phosphate hydrolysis, however, our data show that IP1 accumulation does not necessarily result in iCa2+ mobilization via mGlu5. It is possible that these differences between IP1 and iCa2+ are related to the different kinetics of response coupling measurements for these two assays. A lack of equilibrium being achieved by allosteric ligands relative to orthosteric agonists in the transient iCa2+ assay may also be a contributing factor. Indeed, underlying temporal differences between assays and ligands influences bias at the dopamine D2 receptor, such that kinetic context may also play a crucial role in understanding biased agonism (Klein Herenbrink et al., 2016). However, it is widely accepted that the mGlu5 receptor is coupled to various calcium channels (Gao et al., 2013; Kammermeier et al., 2000; Lu et al., 1999; McCool et al., 1998; Ribeiro et al., 2010a; Tu et al., 1999), thereby resulting in Ca2+ mobilization from both intracellular and extracellular stores. In order to differentiate the sources of Ca2+, we compared iCa2+ mobilization in the presence and absence of 1.2 mM Ca2+ in HEK293A-mGlu5-low cells. Interestingly, while glutamate and DHPG had reduced maximal responses in the absence of extracellular Ca2+, all studied mGlu5 allosteric ligands were unaffected. By applying a rigorous analytical approach to quantify bias, we found that VU0424465 showed significant bias between IP1 and extracellular Ca2+-free iCa2+ assays (20-fold) and between IP1 and iCa2+ assays (110-fold). These data suggest that allosteric agonists couple to a different complement of effectors than orthosteric agonists to influence intracellular Ca2+ levels.
Our data clearly demonstrate that allosteric modulator activity in an mGlu5-iCa2+ mobilization assay is not necessarily predictive of activity for other mGlu5-mediated effects. Our data contribute to a growing body of evidence for biased agonism at metabotropic glutamate receptors including for mGlu1 (Emery et al., 2012; Hathaway et al., 2015) and group III mGlu receptors (Jalan-Sakrikar et al., 2014). Further, we observed probe-dependence for potentiation of mGlu5-iCa2+ mobilization in HEK293A cells, where the magnitude of cooperativity for select allosteric ligands was dependent upon the orthosteric agonist present. Probe dependence has not previously been observed for mGlu5, however, is known to be prevalent for other mGlu subtypes (Jalan-Sakrikar et al, 2014). The phenomenon of probe dependence is a crucial consideration, particularly when translating findings from in vitro assays to native preparations, such as slice electrophysiology, where surrogate agonists are often used. Interestingly, while VU29 had higher positive cooperativity with DHPG compared to glutamate, CDPPB potentiated both agonists to a similar extent. Similarly, for the three picolinamide acetylenes studied, VU0424465 and VU0405398 had differential cooperativities with DHPG and glutamate whereas VU0403602 did not. These data demonstrate that the probe dependence observed is not linked to a particular chemotype, or consistent between ligands from the same scaffold.
While probe dependence is an important consideration when moving from recombinant to physiologically relevant systems, the change in cell background in itself may be a confounding factor in understanding drug pharmacology. Indeed, in this study, we observed clear differences in the temporal profile for mGlu5-ERK1/2 phosphorylation between HEK293A-mGlu5-low and cortical neurons. While ERK1/2 phosphorylation was transient in HEK293A-mGlu5-low cells, in cortical neurons pERK1/2 levels remained elevated over 30min. These data suggest that distinct intracellular effectors are mediating pERK1/2 in response to mGlu5 activation in cortical neurons versus HEK293A cells. Moreover, sustained ligand-induced responses suggest that the consequences of receptor activation will continue after a ligand has vacated the receptor. It is also possible that cellular context is driving differential compartmentalization and/or internalization of mGlu5 in these two cell types. Of note, 50–80% of mGlu5 receptors are thought to be located on intracellular membranes (Hubert et al., 2001; Kumar et al., 2008), and subcellular localisation of mGlu5 can dictate the overall cellular response (Jong et al., 2009; Kumar et al., 2012). It remains to be determined whether or not the mGlu5 allosteric ligands tested cross the plasma membrane, induce receptor internalization or are internalized with the receptor. DHPG is not actively transported across the plasma membrane, only activating receptors at the cell surface (Jong et al., 2009), this suggests that the sustained pERK1/2 response in cortical neurons originates from cell surface receptors, however, it is possible that the continued signaling may be driven by internalised cell surface receptors within endosomes.
Comparison of the biased agonism signaling fingerprints for all eight allosteric ligands revealed that VU0424465 had the most divergent profile in both HEK cells and neuronal cultures. Our earlier study linked VU0424465 intrinsic efficacy (for mGlu5-iCa2+ in HEK cells) to seizure activity and behavioral convulsions (Rook et al., 2013). However, comparison of the bias signaling fingerprint of VU0424465 with that of DPFE, VU0409551 and CDPPB, three ligands that have efficacy in models of cognition and psychosis but no reported adverse effects (Gregory et al., 2013a; Uslaner et al., 2009; Balu et al., 2016; Rook et al., 2015), suggests that the pronounced bias (10–100 fold) of VU0424465 toward IP1 and ERK1/2 relative to iCa2+ in HEK293A-mGlu5-low cells may be a better prediction of adverse effect liability. It is worth noting that all experiments in cortical neurons were performed in the presence of CPCCOEt (an mGlu1 negative allosteric modulator) to eliminate DHPG activation of mGlu1. However, mGlu1 and mGlu5 are known to heteromerize (Sevastyanova and Kammermeier, 2014). Heteromerization of metabotropic glutamate receptors (mGlu2 and mGlu4) can influence allosteric modulator pharmacology (Yin et al., 2014). In the future it would be of interest to determine if mGlu1/mGlu5 heteromers influence the biased agonism observed for mGlu5 selective PAM-agonists. Understanding the bias profile of different ligands, and linking this to known preclinical profiles, offers the opportunity to design therapeutics that avoid signaling pathways associated with adverse effects.
We found that allosteric ligand bias profiles differed between HEK293A-mGlu5-low cells and cortical neuronal cultures. Of note, VU0409551 was the only ligand to show significant bias away from pERK1/2 (relative to IP1) and a lack of agonist efficacy for iCa2+ mobilization in cortical neurons. These data are consistent with the previous report that VU0409551 acting at mGlu5 does not potentiate NMDA channel currents (Rook et al., 2015) or stimulate pERK1/2 levels in the hippocampus or prefrontal cortex when dosed chronically (Balu et al., 2016). Interestingly, DPFE and VU0405398 had distinctly different concentration-response relationships for ERK1/2 phosphorylation in cortical neurons, eliciting a bi-phasic response. While VU0405398 has not yet been tested in vivo, DPFE has a unique preclinical efficacy profile. Considerably lower doses of DPFE (0.56mg/kg) are required for cognition enhancement, whereas >10mg/kg is needed for efficacy in reversing amphetamine-induced hyperlocomotion (Gregory et al., 2013). In contrast, cognition studies with CDPPB (Horio et al., 2013; Stefani and Moghaddam, 2010) have demonstrated efficacy at doses similar those required for efficacy in reversing amphetamine-induced hyperlocomotion (Kinney et al., 2005). It is tempting to speculate that the distinct coupling profiles for DPFE and VU0409551 with respect to pERK1/2 may be predictive of in vivo efficacy that is biased toward cognition enhancement.
In addition to biased agonism, biased modulation was also demonstrated for different mGlu5 allosteric ligands in cortical neurons. Biased modulation may be more favorable for CNS drugs, rather than direct activation of receptors, as it allows for greater control over basal glutamatergic tone. Multiple studies have demonstrated that different mGlu5 effectors and second messengers can be perturbed in different CNS disorders. For example, disrupted mTOR and Akt signaling is associated with major depressive disorder and suicide (Dwivedi et al., 2010; Jernigan et al., 2011) whereas increases in basal pERK1/2, Akt and altered IP3/iCa2+ signaling are observed in a mouse model of Huntington’s disease (Bezprozvanny and Hayden, 2004; Ribeiro et al., 2010b; Tang et al., 2005). Therefore, biased modulation may provide the means to tailor enhancement or inhibition of mGlu5 activity to effectors associated with different disease states.
In summary, we have determined the signaling fingerprint of several mGlu5 allosteric ligands at iCa2+ mobilization, IP1 accumulation and ERK1/2 phosphorylation in both recombinant and native systems. Probe dependence, biased agonism and biased modulation were operative at this receptor. Importantly, the most divergent bias profiles were associated with ligands previously shown to possess distinct in vivo efficacy profiles. Drug discovery for neuroscience-related disorders continues to suffer from high attrition rates, in large part due to an inability to translate promising preclinical pharmacological profiles to clinically effective therapeutics. Our findings suggest that unappreciated biased agonism and/or modulation may contribute to these failures and highlight the need to examine multiple measures of receptor activity to best classify the pharmacology of ligands. In the future, these biased signaling fingerprints may provide a framework that can be used to rationally develop mGlu5 allosteric ligands with optimal in vitro profiles that translate to desirable activity in preclinical models and avoid on-target adverse effects.
Supplementary Material
Highlights.
mGlu5 enhancers differentially affect signaling and are biased agonists/modulators
Biased mGlu5 agonism is evident in both recombinant cells and neuronal cultures
Differences noted in vivo may be due to biased mGlu5 allosteric agonism/modulation
Acknowledgments
This work was supported by the National Health & Medical Research Council of Australia (NHMRC): CJ Martin Overseas Biomedical postdoctoral training Fellowship (KJG: APP1013709), Project Grant APP1084775 (KJG), Program Grant APP1055134 (AC) and Senior Principal Research Fellowship APP1102950 (AC). Work within the Vanderbilt Center for Neuroscience Drug Discovery on mGlu5 PAMs was supported by the NIH/NIMH R01MH062646. PJC is an inventor on patents that protect multiple classes of mGlu5 PAMs and receives research support from AstraZeneca. ERK SureFire kits were a kind gift from Dr. Michael Crouch and TGR Biosciences (Thebarton, Australia). We thank Ms. Sabine Albold for expert technical assistance.
Abbreviations
- cAMP
cyclic adenosine monophosphate
- CDPPB
3-Cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide
- CNS
central nervous system
- CPCCOEt
7-(hydroxyimino)cyclopropa[b] chromen-1a-carboxylate ethyl ester
- DHPG
(S)-3,5-dihydroxyphenylglycine
- DMEM
Dulbecco’s modified Eagle’s medium
- DPFE
1-(4-(2,4-difluorophenyl)piperazin-1-yl)-2-((4-fluorobenzyl)oxy)ethanone
- eCa2+
extracellular calcium
- ERK1/2
extracellular signal-regulated kinases 1 and 2
- FBS
fetal bovine serum
- GPCR
G protein-coupled receptor
- GPT
glutamic pyruvic transaminase
- HBSS
Hank’s Balanced Salt Solution
- HEK293A
human embryonic kidney 293
- iCa2+
intracellular calcium
- IP1
inositol 1-phosphate
- mGlu5
metabotropic glutamate receptor subtype 5
- MPEP
2-Methyl-6-(phenylethynyl)pyridine
- mTOR
mammalian target of rapamycin
- NAL
neutral allosteric ligand
- NAM
negative allosteric modulator
- Opti-MEM
Opti- modified Eagle’s medium
- PAM
positive allosteric modulator
- PI3K
phosphoinositide 3-kinase
- VU0360172
N-cyclobutyl-6-((3-fluorophenyl)ethynyl)picolinamide
- VU0403602
N-cyclobutyl-5-((3-fluorophenyl)ethynyl)picolinamide
- VU0424465
(R)-5-((3-fluorophenyl)ethynyl)-N-(3-hydroxy-3-methylbutan-2-yl)picolinamide
- VU0405398
(5-((3-fluorophenyl)ethynyl)pyridin-2-yl)(3-hydroxyazetidin-1-yl)methanone
- VU0409551
((4-fluorophenyl)(2-(phenoxymethyl)-6,7-dihydrooxazolo[5,4-c]pyridin-5(4H)-yl)methanone)
- VU29
N-(1,3-diphenyl-1H-pyrazolo-5-yl)-4-nitrobenzamide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ayala JE, Chen Y, Banko JL, Sheffler DJ, Williams R, Telk AN, Watson NL, Xiang Z, Zhang Y, Jones PJ, Lindsley CW, Olive MF, Conn PJ. mGluR5 positive allosteric modulators facilitate both hippocampal LTP and LTD and enhance spatial learning. Neuropsychopharmacol. 2009;34:2057–2071. doi: 10.1038/npp.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balu DT, Li Y, Takagi S, Presti KT, Ramikie TS, Rook JM, Jones CK, Lindsley CW, Conn PJ, Bolshakov VY, Coyle JT. An mGlu5-Positive Allosteric Modulator Rescues the Neuroplasticity Deficits in a Genetic Model of NMDA Receptor Hypofunction in Schizophrenia. Neuropsychopharmacol. 2016;41:2052–2061. doi: 10.1038/npp.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezprozvanny I, Hayden MR. Deranged neuronal calcium signaling and Huntington disease. Biochem Biophys Res Commun. 2004;322:1310–1317. doi: 10.1016/j.bbrc.2004.08.035. [DOI] [PubMed] [Google Scholar]
- Bridges TM, Rook JM, Noetzel MJ, Morrison RD, Zhou Y, Gogliotti RD, Vinson PN, Xiang Z, Jones CK, Niswender CM, Lindsley CW, Stauffer SR, Conn PJ, Daniels JS. Biotransformation of a novel positive allosteric modulator of metabotropic glutamate receptor subtype 5 contributes to seizure-like adverse events in rats involving a receptor agonism-dependent mechanism. Drug Metab Dispos. 2013;41:1703–1714. doi: 10.1124/dmd.113.052084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Nong Y, Goudet C, Hemstapat K, de Paulis T, Pin JP, Conn PJ. Interaction of novel positive allosteric modulators of metabotropic glutamate receptor 5 with the negative allosteric antagonist site is required for potentiation of receptor responses. Mol Pharmacol. 2007;71:1389–1398. doi: 10.1124/mol.106.032425. [DOI] [PubMed] [Google Scholar]
- Christopoulos A, Kenakin T. G protein-coupled receptor allosterism and complexing. Pharmacol Rev. 2002;54:323–374. doi: 10.1124/pr.54.2.323. [DOI] [PubMed] [Google Scholar]
- Cleva RM, Hicks MP, Gass JT, Wischerath KC, Plasters ET, Widholm JJ, Olive MF. mGluR5 positive allosteric modulation enhances extinction learning following cocaine self-administration. Behav Neurosci. 2011;125:10–19. doi: 10.1037/a0022339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde-Ceide S, Martinez-Viturro CM, Alcazar J, Garcia-Barrantes PM, Lavreysen H, Mackie C, Vinson PN, Rook JM, Bridges TM, Daniels JS, Megens A, Langlois X, Drinkenburg WH, Ahnaou A, Niswender CM, Jones CK, Macdonald GJ, Steckler T, Conn PJ, Stauffer SR, Bartolome-Nebreda JM, Lindsley CW. Discovery of VU0409551/JNJ-46778212: An mGlu5 Positive Allosteric Modulator Clinical Candidate Targeting Schizophrenia. ACS Med Chem Lett. 2015;6:716–720. doi: 10.1021/acsmedchemlett.5b00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore AS, Okrasa K, Patel JC, Serrano-Vega M, Bennett K, Cooke RM, Errey JC, Jazayeri A, Khan S, Tehan B, Weir M, Wiggin GR, Marshall FH. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature. 2014;511:557–562. doi: 10.1038/nature13396. [DOI] [PubMed] [Google Scholar]
- Doria JG, de Souza JM, Andrade JN, Rodrigues HA, Guimaraes IM, Carvalho TG, Guatimosim C, Dobransky T, Ribeiro FM. The mGluR5 positive allosteric modulator, CDPPB, ameliorates pathology and phenotypic signs of a mouse model of Huntington’s disease. Neurobiol Dis. 2015;73:163–173. doi: 10.1016/j.nbd.2014.08.021. [DOI] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Zhang H, Roberts RC, Conley RR, Pandey GN. Modulation in activation and expression of phosphatase and tensin homolog on chromosome ten, Akt1, and 3-phosphoinositide-dependent kinase 1: further evidence demonstrating altered phosphoinositide 3-kinase signaling in postmortem brain of suicide subjects. Biol Psychiat. 2010;67:1017–1025. doi: 10.1016/j.biopsych.2009.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery AC, Pshenichkin S, Takoudjou GR, Grajkowska E, Wolfe BB, Wroblewski JT. The protective signaling of metabotropic glutamate receptor 1 Is mediated by sustained, beta-arrestin-1-dependent ERK phosphorylation. J Biol Chem. 2010;285:26041–26048. doi: 10.1074/jbc.M110.139899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery AC, DiRaddo JO, Miller E, Hathaway HA, Pshenichkin S, Takoudjou GR, Grajkowska E, Yasuda RP, Wolfe BB, Wroblewski JT. Ligand bias at metabotropic glutamate 1a receptors: molecular determinants that distinguish beta-arrestin-mediated from G protein-mediated signaling. Mol Pharmacol. 2012;82:291–301. doi: 10.1124/mol.112.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganella DE, Thangaraju P, Lawrence AJ, Kim JH. Fear extinction in 17 day old rats is dependent on metabotropic glutamate receptor 5 signaling. Behav Brain Res. 2014;298:32–36. doi: 10.1016/j.bbr.2014.12.010. [DOI] [PubMed] [Google Scholar]
- Gao C, Tronson NC, Radulovic J. Modulation of behavior by scaffolding proteins of the post-synaptic density. Neurobiol Learn Mem. 2013;105:3–12. doi: 10.1016/j.nlm.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass JT, Trantham-Davidson H, Kassab AS, Glen WB, Jr, Olive MF, Chandler LJ. Enhancement of extinction learning attenuates ethanol-seeking behavior and alters plasticity in the prefrontal cortex. J Neurosce. 2014;34:7562–7574. doi: 10.1523/JNEUROSCI.5616-12.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry PR, Sexton PM, Christopoulos A. Novel Allosteric Modulators of G Protein-coupled Receptors. J Biol Chem. 2015;290:19478–19488. doi: 10.1074/jbc.R115.662759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory KJ, Herman EJ, Ramsey AJ, Hammond AS, Byun NE, Stauffer SR, Manka JT, Jadhav S, Bridges TM, Weaver CD, Niswender CM, Steckler T, Drinkenburg WH, Ahnaou A, Lavreysen H, Macdonald GJ, Bartolome JM, Mackie C, Hrupka BJ, Caron MG, Daigle TL, Lindsley CW, Conn PJ, Jones CK. N-aryl piperazine metabotropic glutamate receptor 5 positive allosteric modulators possess efficacy in preclinical models of NMDA hypofunction and cognitive enhancement. J Pharmacol Exp Ther. 2013a;347:438–457. doi: 10.1124/jpet.113.206623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory KJ, Nguyen ED, Malosh C, Mendenhall JL, Zic JZ, Bates BS, Noetzel MJ, Squire EF, Turner EM, Rook JM, Emmitte KA, Stauffer SR, Lindsley CW, Meiler J, Conn PJ. Identification of specific ligand-receptor interactions that govern binding and cooperativity of diverse modulators to a common metabotropic glutamate receptor 5 allosteric site. ACS Chem Neurosci. 2014;5:282–295. doi: 10.1021/cn400225x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory KJ, Nguyen ED, Reiff SD, Squire EF, Stauffer SR, Lindsley CW, Meiler J, Conn PJ. Probing the metabotropic glutamate receptor 5 (mGlu5) positive allosteric modulator (PAM) binding pocket: discovery of point mutations that engender a “molecular switch” in PAM pharmacology. Mol Pharmacol. 2013b;83:991–1006. doi: 10.1124/mol.112.083949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory KJ, Noetzel MJ, Niswender CM. Pharmacology of metabotropic glutamate receptor allosteric modulators: structural basis and therapeutic potential for CNS disorders. Prog Mol Biol Translat Sci. 2013c;115:61–121. doi: 10.1016/B978-0-12-394587-7.00002-6. [DOI] [PubMed] [Google Scholar]
- Gregory KJ, Noetzel MJ, Rook JM, Vinson PN, Stauffer SR, Rodriguez AL, Emmitte KA, Zhou Y, Chun AC, Felts AS, Chauder BA, Lindsley CW, Niswender CM, Conn PJ. Investigating metabotropic glutamate receptor 5 allosteric modulator cooperativity, affinity, and agonism: enriching structure-function studies and structure-activity relationships. Mol Pharmacol. 2012;82:860–875. doi: 10.1124/mol.112.080531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathaway HA, Pshenichkin S, Grajkowska E, Gelb T, Emery AC, Wolfe BB, Wroblewski JT. Pharmacological characterization of mGlu1 receptors in cerebellar granule cells reveals biased agonism. Neuropharmacol. 2015;93:199–208. doi: 10.1016/j.neuropharm.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio M, Fujita Y, Hashimoto K. Therapeutic effects of metabotropic glutamate receptor 5 positive allosteric modulator CDPPB on phencyclidine-induced cognitive deficits in mice. Fundam Clin Pharmacol. 2013;27:483–488. doi: 10.1111/j.1472-8206.2012.01045.x. [DOI] [PubMed] [Google Scholar]
- Hubert GW, Paquet M, Smith Y. Differential subcellular localization of mGluR1a and mGluR5 in the rat and monkey Substantia nigra. Journal of Neuroscience. 2001;21:1838–1847. doi: 10.1523/JNEUROSCI.21-06-01838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernigan CS, Goswami DB, Austin MC, Iyo AH, Chandran A, Stockmeier CA, Karolewicz B. The mTOR signaling pathway in the prefrontal cortex is compromised in major depressive disorder. Prog Neuro-Psychoph. 2011;35:1774–1779. doi: 10.1016/j.pnpbp.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalan-Sakrikar N, Field JR, Klar R, Mattmann ME, Gregory KJ, Zamorano R, Engers DW, Bollinger SR, Weaver CD, Days EL, Lewis LM, Utley TJ, Hurtado M, Rigault D, Acher F, Walker AG, Melancon BJ, Wood MR, Lindsley CW, Conn PJ, Xiang Z, Hopkins CR, Niswender CM. Identification of positive allosteric modulators VU0155094 (ML397) and VU0422288 (ML396) reveals new insights into the biology of metabotropic glutamate receptor 7. ACS Chem Neurosci. 2014;5:1221–1237. doi: 10.1021/cn500153z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jong YJ, Kumar V, O’Malley KL. Intracellular metabotropic glutamate receptor 5 (mGluR5) activates signaling cascades distinct from cell surface counterparts. J Biol Chem. 2009;284:35827–35838. doi: 10.1074/jbc.M109.046276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammermeier PJ, Xiao B, Tu JC, Worley PF, Ikeda SR. Homer proteins regulate coupling of group I metabotropic glutamate receptors to N-type calcium and M-type potassium channels. J Neurosci. 2000;20:7238–7245. doi: 10.1523/JNEUROSCI.20-19-07238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin T, Christopoulos A. Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat Rev Drug Discov. 2013;12:205–216. doi: 10.1038/nrd3954. [DOI] [PubMed] [Google Scholar]
- Kenakin T, Watson C, Muniz-Medina V, Christopoulos A, Novick S. A simple method for quantifying functional selectivity and agonist bias. ACS Chem Neurosci. 2012;3:193–203. doi: 10.1021/cn200111m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney GG, O’Brien JA, Lemaire W, Burno M, Bickel DJ, Clements MK, Chen TB, Wisnoski DD, Lindsley CW, Tiller PR, Smith S, Jacobson MA, Sur C, Duggan ME, Pettibone DJ, Conn PJ, Williams DL., Jr A novel selective positive allosteric modulator of metabotropic glutamate receptor subtype 5 has in vivo activity and antipsychotic-like effects in rat behavioral models. J Pharmacol Exp Ther. 2005;313:199–206. doi: 10.1124/jpet.104.079244. [DOI] [PubMed] [Google Scholar]
- Klein Herenbrink C, Sykes DA, Donthamsetti P, Canals M, Coudrat T, Shonberg J, Scammells PJ, Capuano B, Sexton PM, Charlton SJ, Javitch JA, Christopoulos A, Lane JR. The role of kinetic context in apparent biased agonism at GPCRs. Nat Commun. 2016;7:10842. doi: 10.1038/ncomms10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Jong YJ, O’Malley KL. Activated nuclear metabotropic glutamate receptor mGlu5 couples to nuclear Gq/11 proteins to generate inositol 1,4,5-trisphosphate-mediated nuclear Ca2+ release. Journal of Biological Chemistry. 2008;283:14072–14083. doi: 10.1074/jbc.M708551200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach K, Sexton PM, Christopoulos A. Allosteric GPCR modulators: taking advantage of permissive receptor pharmacology. Trends Pharmacol Sci. 2007;28:382–389. doi: 10.1016/j.tips.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Lindsley CW, Wisnoski DD, Leister WH, O’Brien JA, Lemaire W, Williams DL, Jr, Burno M, Sur C, Kinney GG, Pettibone DJ, Tiller PR, Smith S, Duggan ME, Hartman GD, Conn PJ, Huff JR. Discovery of positive allosteric modulators for the metabotropic glutamate receptor subtype 5 from a series of N-(1,3-diphenyl-1H- pyrazol-5-yl)benzamides that potentiate receptor function in vivo. J Med Chem. 2004;47:5825–5828. doi: 10.1021/jm049400d. [DOI] [PubMed] [Google Scholar]
- Lu WY, Xiong ZG, Lei S, Orser BA, Dudek E, Browning MD, MacDonald JF. G-protein-coupled receptors act via protein kinase C and Src to regulate NMDA receptors. Nat Neurosci. 1999;2:331–338. doi: 10.1038/7243. [DOI] [PubMed] [Google Scholar]
- Manka J, Zhou Y, Chun A, Dawson ES, Vinson PN, Niswender CM, Noetzel MJ, Rook JM, Bridges TM, Daniels JS, Jones C, Conn PJ, Lindsley CW, Stauffer SR. Probe Reports from the NIH Molecular Libraries Program. National Center for Biotechnology Information (US); Bethesda (MD): 2010. Identification of a Selective Allosteric Agonist of mGlu5. [PubMed] [Google Scholar]
- McCool BA, Pin JP, Harpold MM, Brust PF, Stauderman KA, Lovinger DM. Rat group I metabotropic glutamate receptors inhibit neuronal Ca2+ channels via multiple signal transduction pathways in HEK 293 cells. J Neurophysiol. 1998;79:379–391. doi: 10.1152/jn.1998.79.1.379. [DOI] [PubMed] [Google Scholar]
- Melancon BJ, Hopkins CR, Wood MR, Emmitte KA, Niswender CM, Christopoulos A, Conn PJ, Lindsley CW. Allosteric modulation of seven transmembrane spanning receptors: theory, practice, and opportunities for central nervous system drug discovery. J Med Chem. 2012;55:1445–1464. doi: 10.1021/jm201139r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswender CM, Conn PJ. Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol. 2010;50:295–322. doi: 10.1146/annurev.pharmtox.011008.145533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noetzel MJ, Gregory KJ, Vinson PN, Manka JT, Stauffer SR, Lindsley CW, Niswender CM, Xiang Z, Conn PJ. A novel metabotropic glutamate receptor 5 positive allosteric modulator acts at a unique site and confers stimulus bias to mGlu5 signaling. Mol Pharmacol. 2013;83:835–847. doi: 10.1124/mol.112.082891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noetzel MJ, Rook JM, Vinson PN, Cho HP, Days E, Zhou Y, Rodriguez AL, Lavreysen H, Stauffer SR, Niswender CM, Xiang Z, Daniels JS, Jones CK, Lindsley CW, Weaver CD, Conn PJ. Functional impact of allosteric agonist activity of selective positive allosteric modulators of metabotropic glutamate receptor subtype 5 in regulating central nervous system function. Mol Pharmacol. 2012;81:120–133. doi: 10.1124/mol.111.075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmentier-Batteur S, Hutson PH, Menzel K, Uslaner JM, Mattson BA, O’Brien JA, Magliaro BC, Forest T, Stump CA, Tynebor RM, Anthony NJ, Tucker TJ, Zhang XF, Gomez R, Huszar SL, Lambeng N, Faure H, Le Poul E, Poli S, Rosahl TW, Rocher JP, Hargreaves R, Williams TM. Mechanism based neurotoxicity of mGlu5 positive allosteric modulators-development challenges for a promising novel antipsychotic target. Neuropharmacol. 2014;82:161–173. doi: 10.1016/j.neuropharm.2012.12.003. [DOI] [PubMed] [Google Scholar]
- Peters J, Scofield MD, Ghee SM, Heinsbroek JA, Reichel CM. Perirhinal Cortex mGlu5 Receptor Activation Reduces Relapse to Methamphetamine Seeking by Restoring Novelty Salience. Neuropsychopharmacol. 2015;41:1477–1485. doi: 10.1038/npp.2015.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro FM, Paquet M, Cregan SP, Ferguson SS. Group I metabotropic glutamate receptor signalling and its implication in neurological disease. CNS Neurol Disord Drug Targets. 2010a;9:574–595. doi: 10.2174/187152710793361612. [DOI] [PubMed] [Google Scholar]
- Ribeiro FM, Paquet M, Ferreira LT, Cregan T, Swan P, Cregan SP, Ferguson SS. Metabotropic glutamate receptor-mediated cell signaling pathways are altered in a mouse model of Huntington’s disease. J Neurosci. 2010b;30:316–324. doi: 10.1523/JNEUROSCI.4974-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AL, Grier MD, Jones CK, Herman EJ, Kane AS, Smith RL, Williams R, Zhou Y, Marlo JE, Days EL, Blatt TN, Jadhav S, Menon UN, Vinson PN, Rook JM, Stauffer SR, Niswender CM, Lindsley CW, Weaver CD, Conn PJ. Discovery of novel allosteric modulators of metabotropic glutamate receptor subtype 5 reveals chemical and functional diversity and in vivo activity in rat behavioral models of anxiolytic and antipsychotic activity. Mol Pharmacol. 2010;78:1105–1123. doi: 10.1124/mol.110.067207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook JM, Noetzel MJ, Pouliot WA, Bridges TM, Vinson PN, Cho HP, Zhou Y, Gogliotti RD, Manka JT, Gregory KJ, Stauffer SR, Dudek FE, Xiang Z, Niswender CM, Daniels JS, Jones CK, Lindsley CW, Conn PJ. Unique signaling profiles of positive allosteric modulators of metabotropic glutamate receptor subtype 5 determine differences in in vivo activity. Biol Psychiatry. 2013;73:501–509. doi: 10.1016/j.biopsych.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook JM, Xiang Z, Lv X, Ghoshal A, Dickerson JW, Bridges TM, Johnson KA, Foster DJ, Gregory KJ, Vinson PN, Thompson AD, Byun N, Collier RL, Bubser M, Nedelcovych MT, Gould RW, Stauffer SR, Daniels JS, Niswender CM, Lavreysen H, Mackie C, Conde-Ceide S, Alcazar J, Bartolome-Nebreda JM, Macdonald GJ, Talpos JC, Steckler T, Jones CK, Lindsley CW, Conn PJ. Biased mGlu5-Positive Allosteric Modulators Provide In Vivo Efficacy without Potentiating mGlu5 Modulation of NMDAR Currents. Neuron. 2015b;86:1029–1040. doi: 10.1016/j.neuron.2015.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengmany K, Gregory KJ. Metabotropic glutamate receptor subtype 5: molecular pharmacology, allosteric modulation and stimulus bias. Br J Pharmacol. 2015 doi: 10.1111/bph.13281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevastyanova TN, Kammermeier PJ. Cooperative signaling between homodimers of metabotropic glutamate receptors 1 and 5. Mol Pharmacol. 2014;86:492–504. doi: 10.1124/mol.114.093468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani MR, Moghaddam B. Activation of type 5 metabotropic glutamate receptors attenuates deficits in cognitive flexibility induced by NMDA receptor blockade. Eur J Pharmacol. 2010;639:26–32. doi: 10.1016/j.ejphar.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TS, Slow E, Lupu V, Stavrovskaya IG, Sugimori M, Llinas R, Kristal BS, Hayden MR, Bezprozvanny I. Disturbed Ca2+ signaling and apoptosis of medium spiny neurons in Huntington’s disease. Proc Natl Acad Sci USA. 2005;102:2602–2607. doi: 10.1073/pnas.0409402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, Worley PF. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–592. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- Uslaner JM, Parmentier-Batteur S, Flick RB, Surles NO, Lam JS, McNaughton CH, Jacobson MA, Hutson PH. Dose-dependent effect of CDPPB, the mGluR5 positive allosteric modulator, on recognition memory is associated with GluR1 and CREB phosphorylation in the prefrontal cortex and hippocampus. Neuropharmacol. 2009;57:531–538. doi: 10.1016/j.neuropharm.2009.07.022. [DOI] [PubMed] [Google Scholar]
- Wu H, Wang C, Gregory KJ, Han GW, Cho HP, Xia Y, Niswender CM, Katritch V, Meiler J, Cherezov V, Conn PJ, Stevens RC. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science. 2014;344:58–64. doi: 10.1126/science.1249489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, Noetzel MJ, Johnson KA, Zamorano R, Jalan-Sakrikar N, Gregory KJ, Conn PJ, Niswender CM. Selective actions of novel allosteric modulators reveal functional heteromers of metabotropic glutamate receptors in the CNS. J Neurosci. 2014;34:79–94. doi: 10.1523/JNEUROSCI.1129-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Rodriguez AL, Conn PJ. Allosteric potentiators of metabotropic glutamate receptor subtype 5 have differential effects on different signaling pathways in cortical astrocytes. J Pharmacol Exp Therap. 2005;315:1212–1219. doi: 10.1124/jpet.105.090308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.