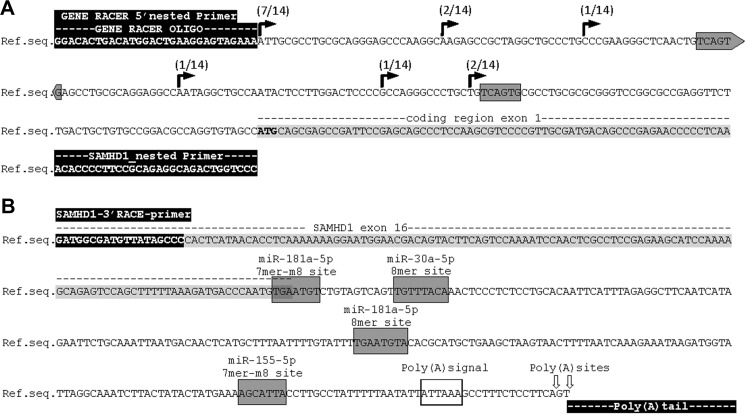
FIGURE 2.
Determination of SAMHD1 transcription start site by 5′-RACE and SAMHD1 3′-UTR by 3′-RACE. A, determination of SAMHD1 transcription start site by 5′-RACE was performed on mRNA from THP-1 and U937 cells. TSS-derived sequences of 5′-RACE of 10 THP-1s and 4 U937s are indicated on the genomic SAMHD1 database sequence NC_000020.11, reference GRCh38.p2 primary assembly. Occurrence of the TSSs in the analyzed data sets is indicated. The SAMHD1 5′-UTR has a length of 69, 82, 112, 152, 173, and 200 nt. GENE RACER 5′ nested primer and SAMHD1 exon 1 primer (SAMHD1_nested primer) are highlighted in black. The coding region of SAMHD1 exon 1 is highlighted in light gray, and the ATG start codon is displayed in boldface letters. Gray boxes mark putative non-canonical offset 6-mer seed regions of miR-181a-3p, as found by seed sequence comparison. B, SAMHD1 3′-UTR was determined by 3′-RACE on mRNA from THP-1 and U937 cells. The SAMHD1 3′-UTR has a total length of 239 nt; in half of the captured sequences, the poly(A)-tail is initiated 2 nt farther downstream, resulting in a 241-nt-long 3′-UTR sequence, as indicated by arrows on the genomic SAMHD1 reference sequence (Ref.seq.) as given in NC_000020.11 reference GRCh38.p2 primary assembly. 3′-RACE sequencing results of nine clones from THP-1 cells and one from U937 cells were analyzed. The SAMHD1 3′-RACE primer is indicated and highlighted in black. The nucleotide sequence belonging to SAMHD1 exon 16 is highlighted in gray. Gray boxes mark canonical seed regions of miR-181a-5p, miR-155–5p, and miR-30a-5p, as found by miRNA target predictions of Miranda, miRcode, TargetScan version 7.1, and seed sequence comparison, and the type of target site is indicated. Poly(A) signal is marked by an open box, poly(A)sites are indicated by arrows, and the poly(A) tail is highlighted in black at the end of the sequence.