FIGURE 7.
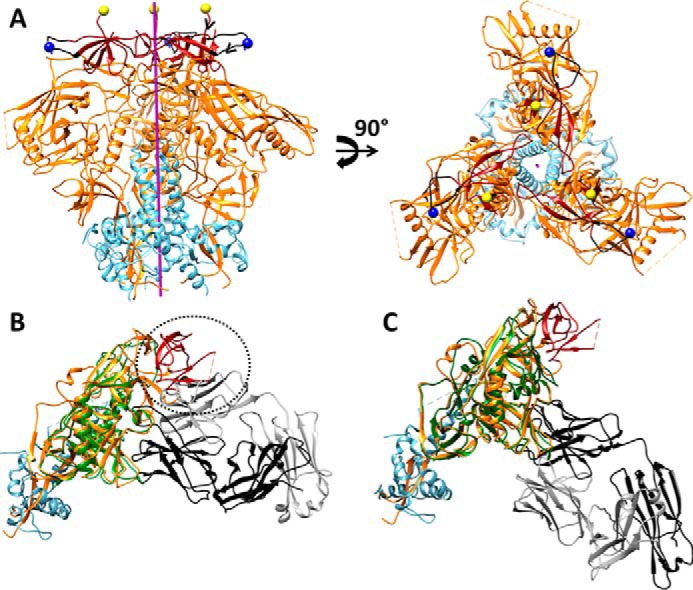
A, approximate positions of new N termini of gp120 in V1cyc and V2cyc, respectively, mapped onto the structure of the BG505 SOSIP.664 HIV-1 Env trimer (Protein Data Bank code 4TVP). The color scheme of the structures is as follows: gp120 (orange), gp41 (cyan), V1V2 region (dark red), residue 141 of V1 region (blue spheres), residue 187 of V2 region (yellow spheres), and region 136–154 of V1 loop (black). The trimer axis is represented as a purple rod. The hCMP trimerization domain was fused to the above N-terminal positions to trimerize gp120. The chain direction at the N termini of V1cyc and V2cyc are indicated by black arrowheads. Residue 144 (the actual N terminus of V1cyc) in JRFL is absent in BG505 because of a deletion of residues 142–149. Hence residue 141 is shown as the N terminus. B, superimposition of core gp120 bound to CD4bs non-neutralizing F105 Fab (PDB code 3HI1) onto gp140 from the BG505 Env trimer (PDB code 4TVP). The color scheme of the structure is as follows: F105 bound gp120 (green), V1V2 region (dark red), and the remaining gp120 subunit from BG505 Env trimer (orange), gp41 from BG505 Env trimer (cyan), F105 Fab light chain (gray), and heavy chain (black) of F105 antigen binding fragment Fab. Inability of F105 to bind gp120 in the presence of the V1V2 region is indicated with a dotted circle demonstrating steric overlap between the F105 light chain and V1V2 in the superimposed structure. C, superimposition of core gp120 bound to CD4bs neutralizing VRC01 Fab (PDB code 3NGB), onto gp140 from the BG505 Env trimer (PDB code 4TVP) shows the absence of any steric clash between the Fab and native trimer. The color scheme of the structure is as follows: VRC01 bound gp120 (green), V1V2 region (dark red), and the gp120 subunit (orange) from BG505 Env trimer, gp41 from BG505 Env trimer (cyan), VRC01 Fab light chain (gray), and heavy chain (black).
