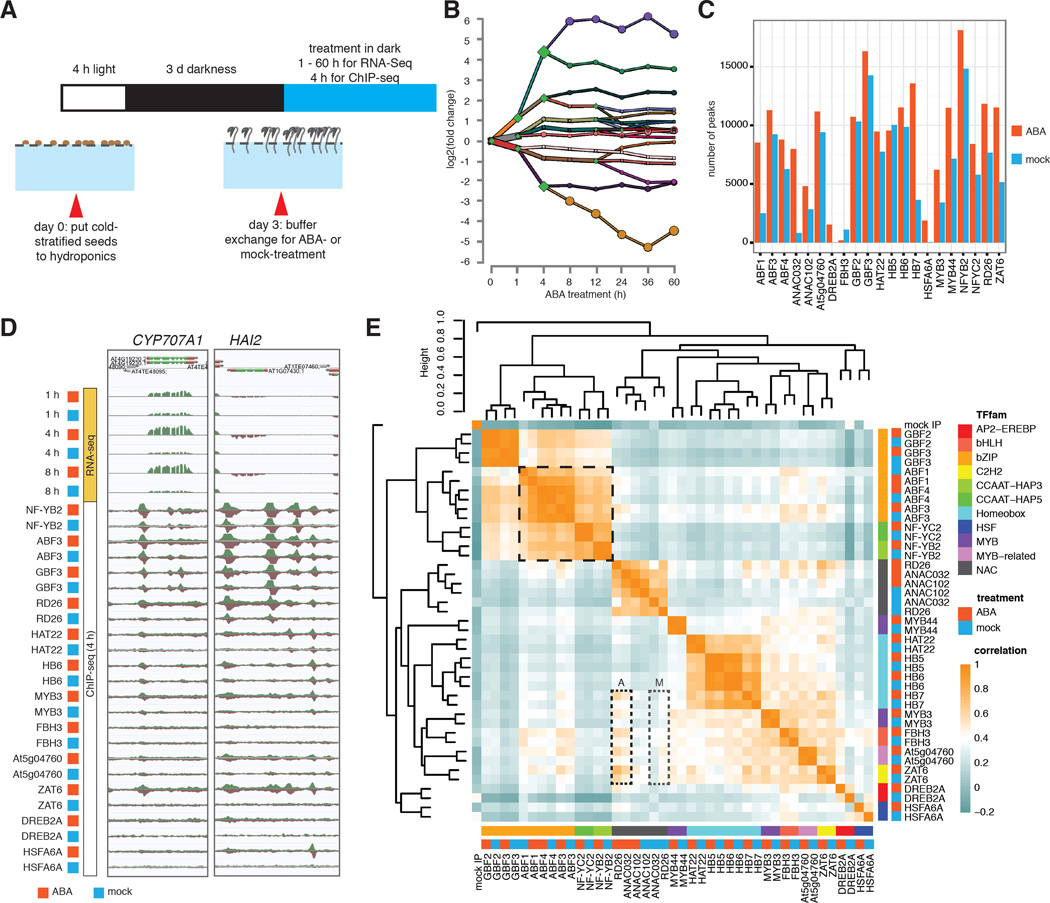
Figure 1. TF identity and hormone treatment determine genome-wide binding profiles.
(A) Growing Arabidopsis thaliana in hydroponics allows convenient buffer exchange for hormone treatment. (B) DREM reconstructed RNA expression paths 60 hours post ABA exposure. Each path corresponds to a set of genes that are co-expressed. Split nodes (green diamonds) represent a temporal event where a group of genes co-expressed up to that point diverge in expression, most likely due to regulatory events. Most splits are observed up to and including the 4h time point, indicating that the majority of regulatory events occur at the first 4 hours. (C) The number of ChIP-seq peaks varies greatly between TFs and treatment conditions. (D) ABA mediated differential gene expression and altered dynamics of TF binding as exemplified by CYP707A1 and HAI2 genes. (E) Comparison of binding correlations based on normalized ChIP-seq read counts near binding sites shows that TFs from same family often have similar binding profiles. TF-TF interaction (bZIPs and NF-Y, black dashed box) and hormone treatment (RD26 and ANAC032, dotted boxes A and M for ABA- and mock-treatment) also contribute to binding profile similarities between TFs.