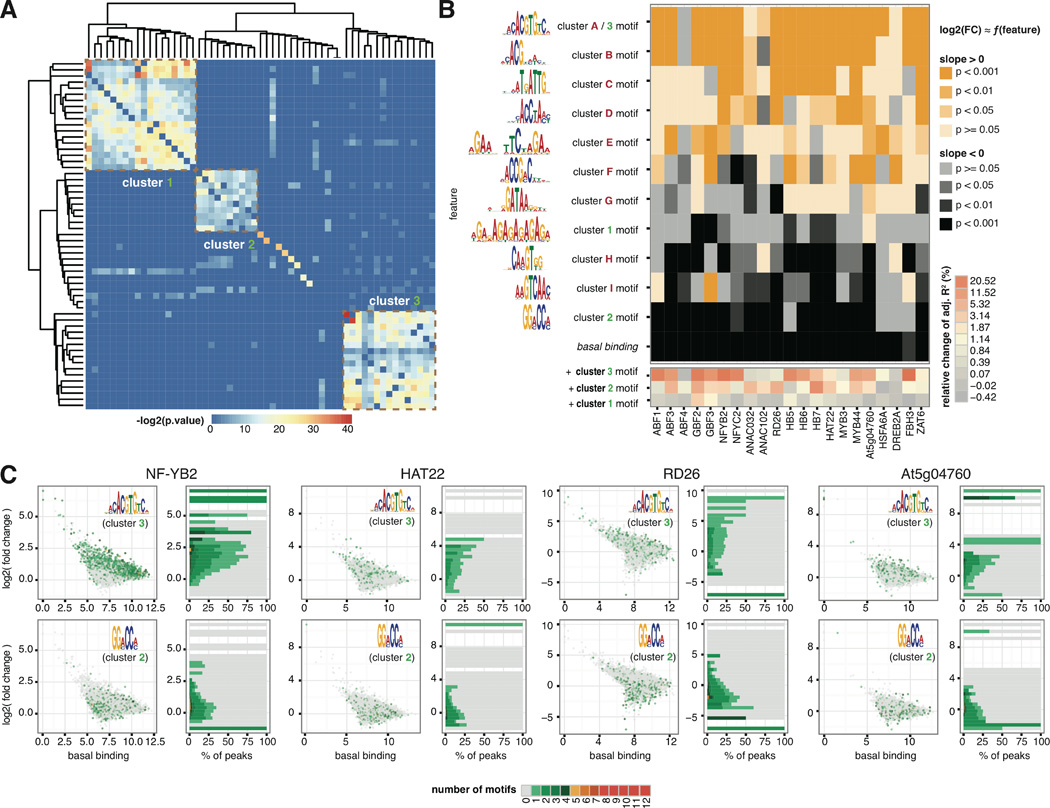
Figure 3. Determinants of TF binding dynamics.
(A) Hierarchical clustering of motifs enriched in dynamic and static peaks revealed three clusters. Each entry in the distance matrix is –log2(p-value) of motif similarity reported by Tomtom (44). (B) Linear regression of differential binding using basal binding and non-redundant sequence features identified positive and negative determinants of dynamic TF binding. Heatmap colors map to two-tailed t-test p-values on the regression coefficients for the null hypothesis that the coefficient is zero. The sequence features were selected from motifs enriched in the strongest peaks in ABA- and mock-treated conditions as well as dynamic and static peaks (Fig. S8). (C) Scatter plots on the left: basal binding of TFs quantified by normalized read count in the peak (x-axis) against log2(fold change) of TF binding after ABA treatment (y-axis), with color of each dot mapped to the number of indicated motifs in the same peak. The occurrence of Cluster 3 and Cluster 2 motifs over the distributions of log2 (fold change) of binding are shown in histograms on the right, with the same color code as the scatter plot. Proportion of peaks containing Cluster 3 motif increases along with log2(fold change) of TF binding for the indicated TFs, whereas proportion of peaks containing Cluster 2 motif are negatively correlated with log2(fold change) of TF binding.