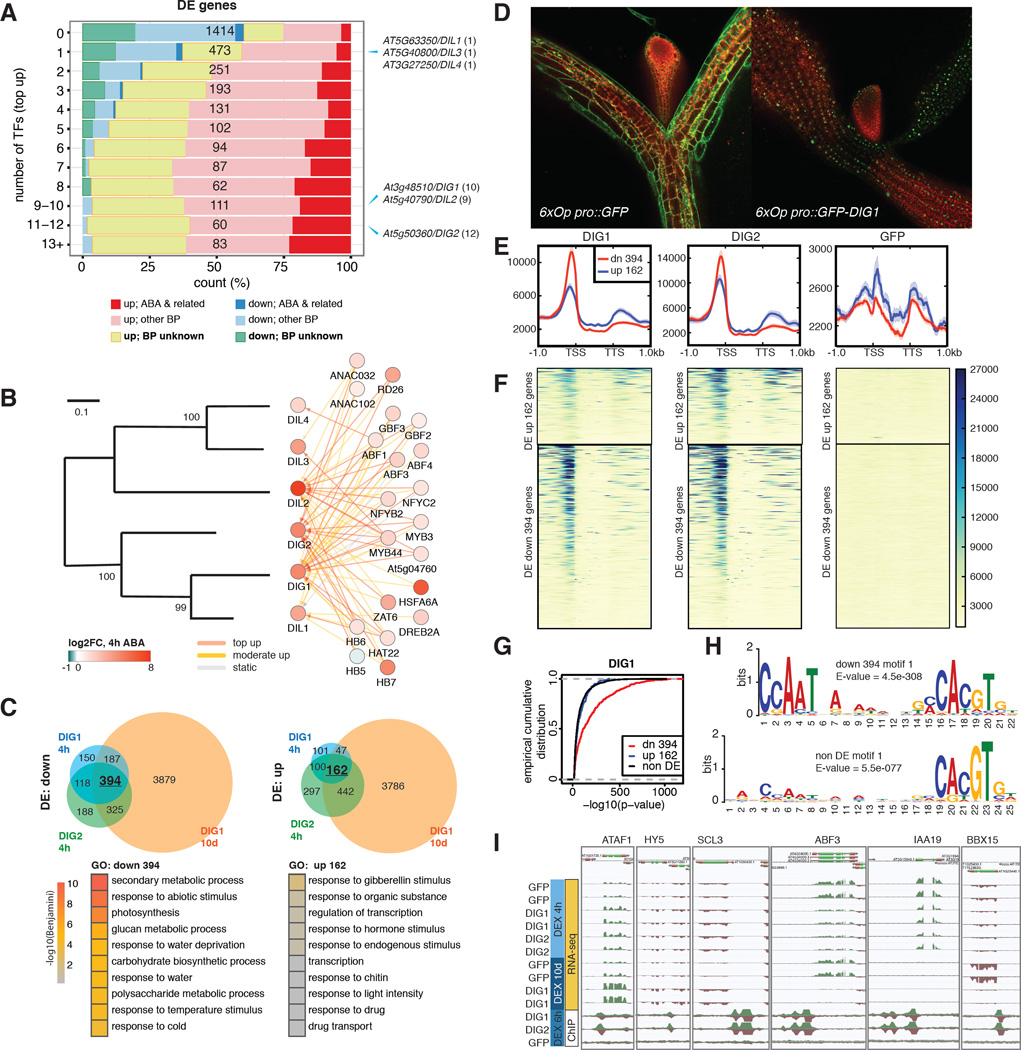
Figure 5. Network analysis identifies new transcriptional regulators of ABA response.
(A) Expression and functional composition of DE grouped by the number of targeting TFs through “top up” binding. Number of genes in each bin is shown in black. The bins to which the DIG/DIL genes belong are indicated on the right, with number of targeting TFs shown in parentheses. (B) DIG/DILs are regulated by multiple ABA-responsive TFs. Left panel: a phylogram of Arabidopsis DIG/DIL proteins. Right Panel: TFs targeting the DIG/DIL genes. (C) DEX-induction of DIGs results in DE of stress- and water-related genes. Upper panels: DE genes by DIGs after indicated period of DEX treatment. Lower panels: top GO terms enriched in DIG DE genes. (D) Confocal imaging of 9-day-old DEX-treated transgenic seedlings shows DIG1 is nuclear localized. (E–F) Metagene profiles (E) and heatmaps (F) of normalized ChIP-seq read counts surrounding DIG DE genes. Down-regulated genes are often associated with strong DIG binding in their promoters. (G) Empirical cumulative distributions of -log10(p-value) of ChIP-seq peaks of DIG1 show it bound more strongly to DIG down-regulated genes than to up-regulated or non-DE genes. (H) A CCAAT(n)8 ABRE motif is strongly enriched near DIG1 binding sites residing within 1 kb of DIG down-regulated genes. Either a weaker motif or no similar motif is enriched in the corresponding regions of non-DE genes or DIG up-regulated genes. (I) Induction of DIGs results in DE of ABA- and developmental-related TFs.