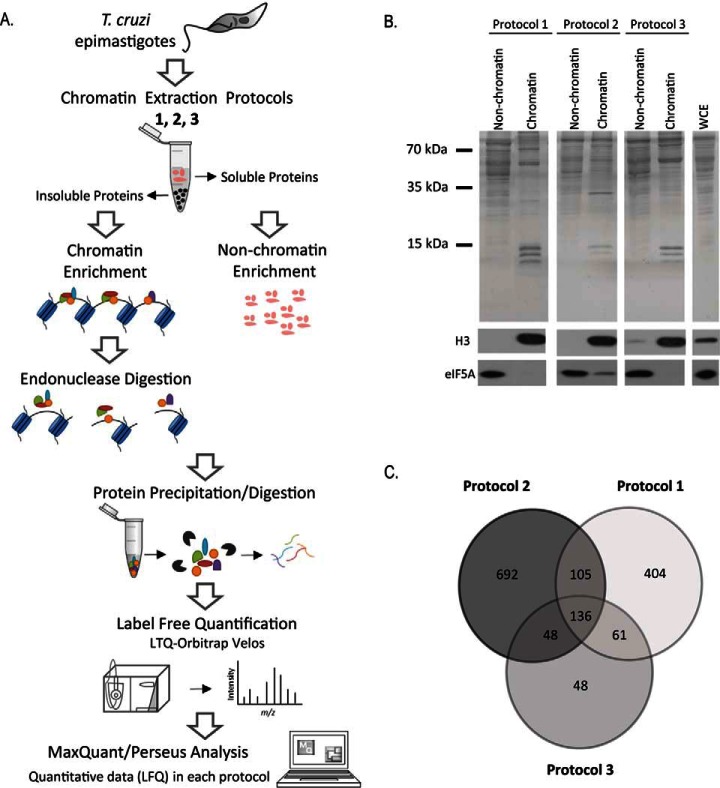
Fig. 1.
A, Outline of the workflow to identify chromatin-associated proteins in T. cruzi. Epimastigotes were harvested and chromatin and nonchromatin enriched extracts were obtained by three different extraction protocols (1, 2 and 3) described at Materials and Methods. After protein precipitation and digestion, peptides were analyzed by LC-MS/MS. Raw data were analyzed by MaxQuant/Perseus software. B, Protein fractionation after chromatin extraction of epimastigotes using three different protocols. Upper panel: SDS-PAGE of nonchromatin and chromatin fractions obtained by protocols 1, 2 and 3. Molecular marker and whole cell extract (WCE) are indicated. Lower panels: Western blotting using antibodies against eIF5A (a nonchromatin protein) and histone H3 (chromatin protein). C, Venn diagram showing the number of proteins found by MS in each chromatin-enrichment protocol.