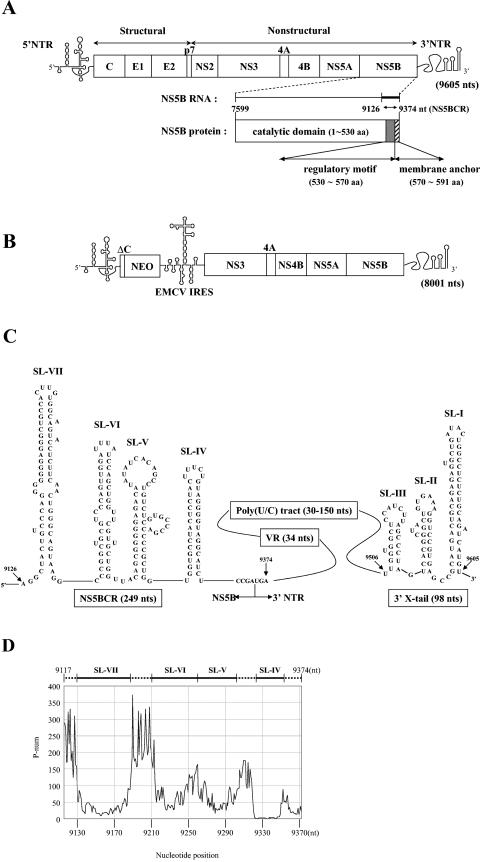
FIG.1.
(A) Genomic structure of full-length HCV RNA. The single-stranded RNA genome of HCV is shown with the 5′ NTR (single line), the structural and nonstructural proteins of the polyprotein (boxed), and the 3′ NTR (single line) indicated. The NS5B domain is shown enlarged both as a coding sequence for the NS5B protein and as an RNA sequence for potential cis-replicating RNA elements. (B) Genomic structure of the subgenomic HCV replicon RNA. The single-stranded RNA genome of the subgenomic replicon contains the HCV 5′ NTR (single line), the neo gene (boxed), the EMCV IRES (single line), the nonstructural proteins of HCV from NS3 to NS5B (boxed), and the 3′ NTR (single line). (C) Predicted RNA stem-loop structures in the HCV 3′ NTR and the C-terminal coding domain of NS5B. Illustrated are the predicted structures of stem-loops IV to VII contained in a 249-nt-long segment of the NS5B coding region (nt 9126 to 9374 of HCV RNA), designated NS5BCR. The single-stranded stretch of nucleotides between stem-loops VI and VII is designated L. Downstream from NS5BCR is the 3′ NTR, which contains a variable region, a poly(U/C) tract, and the 3′ X tail. The highly conserved 3′ X domain is predicted to consist of three stem-loops, I to III (15, 68, 69). (D) P-num values of a 257-nt-long segment (nt 9117 to 9374) of the NS5B C-terminal coding sequence, including all of the NS5BCR sequence, were obtained for HCV-J1 from the website www.virology.wisc.edu/acp/RNAFolds. Stem-loops were identified by consistently low P-num values. aa, amino acids.