Abstract
Influenza B virus contains four integral membrane proteins in its envelope. Of these, BM2 has recently been found to have ion channel activity and is considered to be a functional counterpart to influenza A virus M2, but the role of BM2 in the life cycle of influenza B virus remains unclear. In an effort to explore its function, a number of BM2 mutant viruses were generated by using a reverse genetics technique. The BM2ΔATG mutant virus synthesized BM2 at markedly lower levels but exhibited similar growth to wild-type (wt) virus. In contrast, the BM2 knockout virus, which did not produce BM2, did not grow substantially but was able to grow normally when BM2 was supplemented in trans by host cells expressing BM2. These results indicate that BM2 is a required component for the production of infectious viruses. In the one-step growth cycle, the BM2 knockout virus produced progeny viruses lacking viral ribonucleoprotein complex (vRNP). The inhibited incorporation of vRNP was regained by trans-supplementation of BM2. An immunofluorescence study of virus-infected cells revealed that distribution of hemagglutinin, nucleoprotein, and matrix (M1) protein of the BM2 knockout virus at the apical membrane did not differ from that of wt virus, whereas the sucrose gradient flotation assay revealed that the membrane association of M1 was greatly affected in the absence of BM2, resulting in a decrease of vRNP in membrane fractions. These results strongly suggest that BM2 functions to capture the M1-vRNP complex at the virion budding site during virus assembly.
Influenza A, B, and C viruses are enveloped viruses that assemble at the plasma membrane and bud from infected cells. Both influenza A and B viruses have eight single-stranded negative-sense RNA segments encapsidated with polymerase proteins and nucleoprotein (NP), forming a viral ribonucleoprotein complex (vRNP). The envelope contains three different membrane proteins, hemagglutinin (HA), neuraminidase (NA), and ion channel protein M2 in influenza A virus and NB in influenza B virus (20). The small proteins, M2 and NB, are transmembrane proteins which adopt an N-out C-in orientation (21, 35, 38) and are incorporated into virions as a relatively minor component (3, 37). On the other hand, in the case of influenza A virus, the most abundant product matrix (M1) protein has multiple functions and plays central roles in virus assembly. M1 interacts with vRNP in the nucleus to mediate nuclear export and to prevent nuclear reentry of vRNP (22, 34) and with the cytoplasmic tail of HA and NA on the plasma membrane to form the inner surface of the lipid bilayer of the envelope (1, 7, 18, 40). These interactions are thought to be essential for particle formation and virus release from infected cells.
Although both influenza A and B viruses possess similarities in the coding strategy of genome RNA segments and in the function of viral proteins, there are some differences between these viruses. One of the differences is found in RNA segment 7 of influenza B virus, which encodes the BM2 protein in addition to encoding M1 by bicistronic mRNA (4, 12). The translational strategy of BM2 is quite unique. The initiation codon of BM2 overlaps the termination codon of M1 in an overlapping stop-start pentanucleotide, TAATG, at nucleotides 769 to 773 (15). BM2 synthesis is triggered by the termination of translation of M1, which is encoded in the upstream region of mRNA (15). Because the amino acid sequence of BM2 has been highly conserved for at least 60 years, from B/Lee/40 to B/Nagoya/20/99 (Influenza Sequence Database, http://www.flu.lanl.gov), and BM2 is detected in cells infected with all influenza B viruses examined to date (15, 27), BM2 is thought to play an important role(s) in the life cycle of influenza B virus.
The BM2 protein was previously shown to be a phosphoprotein that is synthesized in the late phase of infection and is incorporated into the virion (27). Recently, Paterson et al. (28) and our group (32) demonstrated by several distinct experimental approaches that BM2 is associated with cellular membranes as an oligomeric integral membrane protein. It was also noted that BM2 accumulated at the Golgi apparatus immediately after synthesis and was then transported to the plasma membrane through the trans-Golgi network and that the N-terminal half of BM2 (residues 2 to 50) was crucial for this transport (32). In addition, Mould et al. (24), by using an electrophysiological technique, indicated that BM2, like influenza A virus M2, has an ion channel activity to permit protons to enter the virions during the uncoating of particles in the endosomes. However, the precise function of BM2 in the influenza B virus life cycle has not yet been fully elucidated. To this end, we generated a selection of influenza B virus BM2 mutants by using a newly constructed reverse genetic system based on the backbone of the B/Yamagata/1/73 (B/Yamagata) virus and examined their replication in tissue culture.
In the present study, we show that BM2 is a necessary component for the generation of infectious influenza B viruses. Furthermore, the dramatic reduction of incorporation of the vRNP complex and M1 into virions in the absence of BM2 suggests that BM2 plays a crucial role in virus assembly of influenza B virus.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Madin-Darby canine kidney (MDCK) cells were grown in Eagle's minimal essential medium supplemented with 10% fetal calf serum (FCS). Human embryonic kidney 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% FCS. Influenza virus was propagated in MDCK cells in Opti-MEM I (Invitrogen) containing 10 μg of trypsin/ml. Polyclonal anti-BM2, anti-NP, anti-M1, and anti-B/Yamagata virus antibodies have been described previously (27). Monoclonal anti-HA antibodies were kindly provided by N. Nakagawa (Institute of Public Health of Kobe City, Hyogo, Japan). Polyclonal anti-β-catenin antibodies and monoclonal anti-β-catenin antibodies were purchased from Sigma-Aldrich.
Plasmid constructs.
The cDNAs of B/Yamagata virus were synthesized by reverse transcription of viral RNA (vRNA) with an oligonucleotide complementary to the conserved 3′ end of the vRNA. The cDNA was amplified by PCR with gene-specific oligonucleotide primers containing the BsmBI or BsaI sites, and PCR products were cloned into a pT7BlueBlunt vector (Novagen) or pUC19. We sequenced at least three independent clones containing all eight RNA segments of B/Yamagata virus and determined a consensus sequence for each gene. After digestion with BsmBI or BsaI, the fragment was subcloned into the BsmBI sites of the PolI plasmid, which contains the human RNA polymerase I promoter and the mouse RNA polymerase I terminator (25). In this report, these constructs for the expression of vRNA are referred to as PolI constructs. The cDNAs encoding the three RNA-dependent RNA polymerase subunits (PB2, PB1, and PA) and NP genes of B/Yamagata virus were cloned into the eukaryotic expression vector pCAGGS/MCS (26). The resulting constructs were designated pCBYPB2, pCBYPB1, pCBYPA, and pCBYNP, which express PB2, PB1, PA, and NP, respectively.
PolI constructs of BM2 knockout mutants and BM2 deletion mutants were constructed as follows. Mutated M genes (Fig. 1) (32) were amplified by PCR from the pT7BlueBlunt plasmid containing the B/Yamagata M gene and were then digested with BsmBI. Primer sequences will be provided upon request. The BsmBI-digested fragment was cloned into the BsmBI sites of the PolI plasmid. The resulting constructs were designated BM2ΔATG, BM2stop, BM2Δ2-23, BM2Δ24-50, BM2Δ51-80, and BM2Δ81-109. All constructs were sequenced to ensure that unwanted mutations were not present.
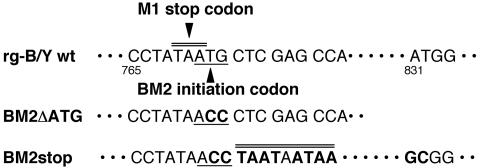
FIG. 1.
Nucleotide sequences of B/Yamagata wt and mutant BM2 constructs. Mutated nucleotides are shown in boldface type. The BM2 initiation codon and the mutated ACC are underlined. Stop codons are indicated by double lines. The numbers shown are nucleotide positions.
The plasmid pCAGGS/BM2, which carried the BM2 gene of B/Yamagata virus, has been described previously (27).
Recovery of infectious virus from cloned DNA.
Transfectant viruses were generated as reported previously (25) with minor modifications. Briefly, 293T cells (0.8 × 106 cells) were plated the day before transfection. Twelve plasmids (8 PolI constructs for 8 RNA segments and 4 protein expression constructs for three polymerase proteins and NP) were mixed with transfection reagent (2 μl of Trans IT LT-1 [Panvera] per μg of DNA) and then added to 293T cells in DMEM-10% FCS. At 16 h posttransfection (hpt), the medium was replaced with Opti-MEM containing 10 μg of trypsin/ml. Forty-eight hours after transfection, viruses in the supernatants were collected.
Indirect immunofluorescence assay (IFA).
MDCK cells grown on coverslips were infected with wild-type (wt) and mutant viruses, fixed, permeabilized, and incubated with the appropriate antibodies as described previously (32). The coverslips were mounted onto glass slides and examined by confocal microscopy (model LSM 510; Carl Zeiss).
Western blotting.
Western blotting analysis was performed as described previously (16). Alternatively, after incubation with the appropriate primary antibody, the membrane was treated with biotinylated goat anti-rabbit antibody and then streptavidin-biotinylated alkaline phosphatase complex. The membrane was developed with 5-bromo-4-chloro-3-indolylphosphate (BCIP) toluidinium-nitroblue tetrazolium substrate (Bio-Rad).
Stable expression of BM2 in MDCK cells.
Hygromycin-resistant MDCK cells stably expressing BM2 (CK/BM2) were established as described previously (23). Briefly, MDCK cells were cotransfected with plasmid pCB7, encoding hygromycin resistance, and plasmid pCAGGS/BM2 at a ratio of 3:1. Stable MDCK cell clones expressing BM2 were selected in medium containing 0.2 mg of hygromycin (Invitrogen)/ml and were screened by IFA. CK/BM2 cells were cultured in DMEM containing 10% FCS.
Real-time quantitative PCR.
RNA was extracted from purified virions by using the Qiagen RNeasy kit according to the manufacturer's instructions. vRNA was converted to cDNA as described above. Primers and probes for the B/Yamagata virus PB1, HA, and M genes were selected by using Primer Express software (Applied Biosystems). The primer and probe sequences for each gene are given below. The probe contains oligonucleotides with the 5′ reporter dye 6-carboxyfluorescein and the 3′ quencher dye 6-carboxytetramethylrhodamine.
Amplification and detection by real-time PCR were performed with the ABI PRISM 7700 sequence detection system (Applied Biosystems). PCR was carried out in 50 μl of reaction mixture consisting of 1× PCR Gold buffer (GeneAmp Gold PCR reagent kit), 250 μM (each) deoxynucleoside triphosphate, 5 mM MgCl2, 1.25 U of AmpliTaq Gold DNA polymerase, 0.2 μM (each) forward and reverse primer, 0.1 μM probe, and 5 μl of cDNA. The reaction conditions were set at 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The standard curve for this assay was calculated by using a series of 10-fold dilutions of PolI plasmids encoding the B/Yamagata virus PB1, HA, and M genes.
The PB1 primers were forward (5′-TGCCAGTAGGTGGAAACGAGA), reverse (5′-TGGTGGGCAGTTACTGAGCA), and probe (5′-AAGGCCAAACTGTCAAATGCAGTGGC). The HA primers were forward (5′-GAAGGAATGATTGCAGGTTGG), reverse (5′-TTAAGGTCTGCTGCCACTGCT), and probe (5′-CGGATACACATCTCATGGAGCACATGGA). The M primers were forward (5′-AGGCGAGAAATGCAAATGGT), reverse (5′-ACGTCTTCTCCCTTCCCCA), and probe (5′-TCAGCTATGAACACAGCAAAAACAATGAATGG).
Flotation analysis.
Flotation analysis was performed as described previously (32). Virus-infected cells were resuspended in 0.5 ml of lysis buffer (10 mM Tris-Cl [pH 7.5], 10 mM KCl, 5 mM MgCl2, and 0.3 μM aprotinin [Roche]) and incubated for 30 min on ice. After incubation, cells were disrupted by repeated passage (50 times) through a 26-gauge needle. Unbroken cells and nuclei were removed by centrifugation at 1,000 × g for 5 min at 4°C. Postnuclear supernatants (0.4 ml) were dispersed into 2 ml of 75% (wt/wt) sucrose in a buffer (TKMB) consisting of 20 mM Tris-Cl (pH 7.5), 25 mM KCl, 5 mM MgCl2, and 0.3 μM aprotinin and placed at the bottom of the tube, then overlaid with 2 ml of 55% (wt/wt) sucrose in TKMB and 0.5 ml of 5% (wt/wt) sucrose in TKMB. The gradient was centrifuged to equilibrium at 150,000 × g for 18 h at 4°C. Fractions (0.5 ml) were collected from the top. Fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Western blotting.
RESULTS
Generation of B/Yamagata and BM2 mutant viruses by reverse genetics with 12 plasmids.
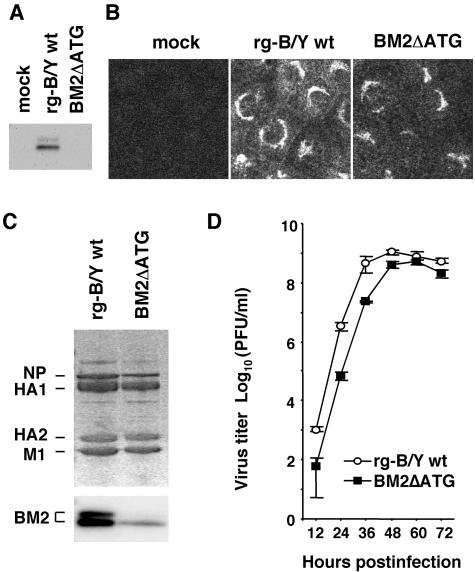
The recently established novel reverse genetics systems allow the generation of infectious influenza A and B viruses with the desired mutations by transfection of either 8 plasmids (13, 14) or 12 to 17 plasmids (9, 17, 25). We adopted a 12-plasmid system for the generation of infectious influenza B viruses. To establish a reverse genetics system for the B/Yamagata virus cDNA backbone, we constructed PolI plasmids that contain cDNAs for the full-length vRNAs of the B/Yamagata virus flanked by the human RNA polymerase I promoter and the mouse RNA polymerase I terminator. These PolI plasmids were cotransfected with four protein expression plasmids (PB1, PB2, PA, and NP) into 293T cells. Using this system, we attempted to generate a BM2 knockout virus designated BM2ΔATG, whose BM2 initiation codon 771ATG was replaced with 771ACC (Fig. 1, middle), and four BM2 deletion mutant viruses, BM2Δ2-23, BM2Δ24-50, BM2Δ51-80, and BM2Δ81-109, whose PolI plasmids contained cDNAs with deletions in the BM2 open reading frame (ORF), as described previously (32). To recover the transfectants, the supernatant of plasmid-transfected 293T cells was harvested at 48 hpt. Table 1 shows the yields of transfectant produced in the supernatant. The BM2ΔATG mutant virus was recovered at similar titers as for the transfectants of B/Yamagata (rg-B/Y) wt virus. In contrast, the four BM2 deletion mutant viruses were not recovered at all. By sequencing RNA segment 7 of the recovered BM2ΔATG mutant virus, we confirmed no reversion from 771ACC to the original 771ATG and no additional mutations in the segment. This result suggests that BM2 synthesis occurs in cells infected with the BM2ΔATG mutant virus despite the absence of the ATG initiation codon in the BM2 ORF. To confirm this, BM2ΔATG mutant virus and rg-B/Y wt virus (control) were infected into MDCK cells and BM2 synthesis was investigated by Western blotting and IFA with anti-BM2 antibody (27). The BM2 signal for the mutant virus was not clearly detected by Western blotting (Fig. 2A), but IFA confirmed weak signals at the Golgi apparatus of infected cells (Fig. 2B). Moreover, extremely small amounts of BM2 were detected in purified virions, and quantitative comparison between the BM2ΔATG mutant and rg-B/Y wt virions indicated that the BM2 content normalized to the HA (HA1 plus HA2) content was 31% of that in rg-B/Y wt virus (Fig. 2C). It should be noted that the ratios of NP to HA of the mutant virus were also decreased slightly compared to those of rg-B/Y wt virus (Fig. 2C).
TABLE 1.
Virus yield of transfectants recovered from 293T cells transfected with 8 poll/BY plasmids and 4 pCA/ORF plasmidsa
| Transfectant virus | Virus yield (PFU/ml) |
|---|---|
| rg-B/Y wt | 1.4 × 104 |
| BM2ΔATG | 3.5 × 104 |
| BM2Δ2-23 | NDb |
| BM2Δ24-50 | ND |
| BM2Δ51-80 | ND |
| BM2Δ81-109 | ND |
293T cells were transfected with 12 plasmids for production of rg-B/Y wt virus or BM2 mutant viruses. The infectious particles in the supernatants were recovered at 48 hpt and titrated in MDCK cells.
ND, not detected.
FIG. 2.
Characterizations of BM2ΔATG mutant virus. (A) MDCK cells were infected with rg-B/Y wt and BM2ΔATG viruses. At 11 hpi, BM2 protein in the cell lysates was detected by Western blotting with anti-BM2 antibody. (B) At 7 hpi, cells were fixed and BM2 was detected with anti-BM2 antibodies by IFA. (C) Protein composition of BM2ΔATG virions. Proteins of purified viruses were analyzed by Coomassie brilliant blue staining (upper panel) and Western blotting with anti-BM2 antibody (lower panel). (D) Multiple-step growth curve of BM2ΔATG mutant viruses. MDCK cells were infected at an MOI of 0.001 PFU, and culture media were harvested at the indicated times postinfection. The virus yield of the supernatant was determined by plaque assay on MDCK cells.
When the multiple-step growth profile of BM2ΔATG mutant virus was compared to that of rg-B/Y wt virus, the mutant virus revealed slightly delayed virus production, but its maximum yield was not significantly different from that of rg-B/Y wt virus (Fig. 2D). Taken together, these results clearly indicate that small amounts of BM2 can facilitate the production of infectious virus.
Rescue of BM2 knockout virus by supplementing BM2 from host cells stably expressing BM2.
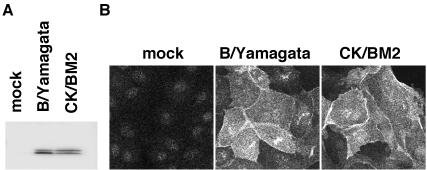
Because BM2ΔATG mutant virus synthesized BM2 at markedly low levels and grew as efficiently as rg-B/Y wt virus, we attempted to produce a genuine BM2 knockout virus. To accomplish this, the PolI plasmid BM2stop, which contained three consecutive stop codons downstream of the 771ACC replacement found in BM2ΔATG and two additional mutations (A831G and T832C), was constructed (Fig. 1, bottom) and transfected into 293T cells in the same manner as for the 12-plasmid system. As expected, no infectious virus was recovered from the supernatant of plasmid-transfected cells (data not shown), thus suggesting that trans-supplementation of BM2 would be necessary for recovery of the mutant. We therefore prepared an MDCK cell line, CK/BM2, that was able to constitutively express BM2 protein, as described in Materials and Methods. In CK/BM2 cells, expression levels and localization of BM2 were similar to those in virus-infected cells at 10 h postinfection (hpi) (Fig. 3).
FIG. 3.
Stable expression of BM2 proteins in MDCK cells. (A) Western blotting analysis with anti-BM2 antibody. Lanes: mock, uninfected cells; B/Yamagata, virus-infected cells; CK/BM2, constitutive BM2-expressing MDCK cell line. (B) Detection of BM2 synthesized in CK/BM2 cells (CK/BM2) at 10 hpi and in virus-infected cells (B/Yamagata) by IFA with the anti-BM2 antibody. Mock, uninfected cells fixed 10 h after incubation.
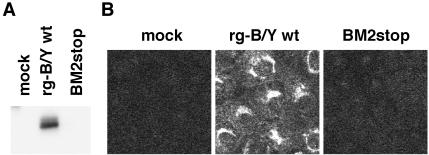
For the rescue of BM2stop mutant virus, 12 plasmids as mentioned above with the BM2 expression plasmid, pCAGGSBM2 (27), were transfected into 293T cells. The supernatant harvested at 48 hpt was then inoculated into CK/BM2 cells to detect transfectants. BM2stop mutant virus was recovered efficiently with a titer similar to that of the rg-B/Y wt and BM2ΔATG viruses (7.6 × 104, 4.4 × 104, and 1.1 × 104 PFU/ml, respectively). To confirm whether rescued BM2stop mutant virus lacked the synthesis of BM2, mutant virus-infected MDCK cells were subjected to Western blotting and IFA. Unlike BM2ΔATG mutant virus, BM2 synthesis was not detected for BM2stop mutant virus in either analysis (Fig. 4), indicating that BM2stop mutant virus was a BM2 knockout virus.
FIG. 4.
Detection of BM2 protein in BM2stop mutant virus-infected MDCK cells. (A) MDCK cells were infected with rg-B/Y wt and BM2stop viruses. At 11 hpi, BM2 protein in the cell lysates was detected by Western blotting with anti-BM2 antibody. (B) At 7 hpi, cells were fixed and BM2 was detected by IFA with anti-BM2 antibodies. mock, uninfected cells.
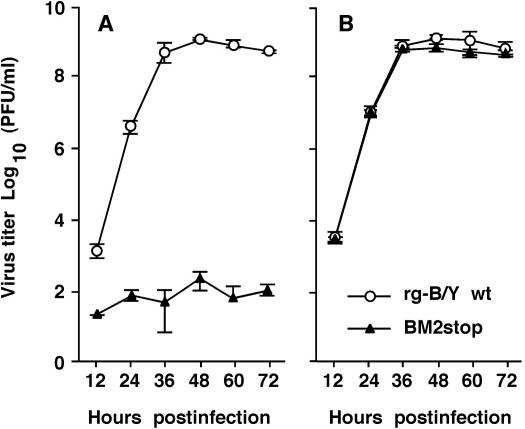
We next compared the kinetics of virus production in multiple-step growth cycles between BM2stop mutant and rg-B/Y wt viruses. In the absence of trans-supplementation of BM2 in MDCK cells, the mutant virus did not grow significantly (Fig. 5A), whereas in the presence of trans-supplementation of BM2 from CK/BM2 cells, the mutant virus regained its ability to grow and exhibited indistinguishable growth kinetics and maximum yield compared to rg-B/Y wt virus (Fig. 5B). These results strongly indicate that BM2 is a necessary component for the production of infectious virus.
FIG. 5.
Growth properties of BM2stop mutant virus. MDCK (A) and CK/BM2 (B) cells were infected at an MOI of 0.001 PFU, and the supernatants of infected cells were harvested at the indicated times postinfection. Virus titers in the supernatant were determined by plaque assay on CK/BM2 cells.
Synthesis and localization of viral proteins in cells infected with BM2 knockout virus.
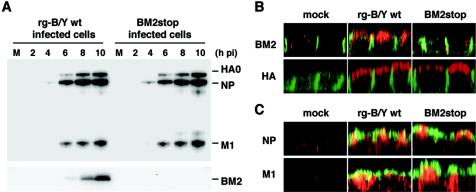
To examine the synthesis and localization of virus proteins in MDCK cells, the cells were infected with BM2stop mutant and rg-B/Y wt viruses. Figure 6A shows the synthesis of viral proteins in MDCK cells every 2 hpi, as detected by Western blotting with anti-BM2 or anti-B/Yamagata virus antibodies (27). BM2 of rg-B/Y wt virus was detected from 6 hpi but was not detected from BM2stop mutant virus throughout infection, which is consistent with the results shown in Fig. 4. On the other hand, the synthesis of major viral proteins, HA, NP, and M1, in the BM2stop mutant virus was similar to that of rg-B/Y wt virus, thus indicating that the synthesis of major viral proteins was not influenced by the deletion of BM2 synthesis.
FIG. 6.
Synthesis and localization of viral proteins in virus-infected MDCK cells. (A) Time course of viral protein synthesis of BM2stop mutant and rg-B/Y wt viruses. Viral proteins were detected by Western blotting with either anti-B/Yamagata (upper panel) or anti-BM2 (lower panel) antibody. Lane M is the lysate of uninfected cells. (B and C) Intracellular localization of the viral proteins of BM2stop mutant and rg-B/Y wt viruses. At 11 hpi, infected cells were fixed and subjected to IFA. Panels show confocal microscopy images of XZ sections, with the apical membrane uppermost. (B) BM2 (red) and HA (red) were detected together with β-catenin (green). β-Catenin is located at the lateral membrane. (C) NP (red) and M1 (red) were detected together with HA (green).
It was previously demonstrated that BM2 is transported to the plasma membrane through the trans-Golgi network (32). Therefore, if BM2 interacted with major viral proteins in the cytoplasm of infected cells, the lack of BM2 may affect the intracellular transport of major viral proteins to the virion budding site at the apical membrane. Accordingly, we observed the distribution of HA, NP, and M1 at the apical membrane in an XZ section by using a confocal microscope. Consistent with the above findings, no BM2 signal was detected for the BM2stop mutant virus at the apical membrane, but BM2 was detected for the rg-B/Y wt virus, as reported by Paterson et al. (28) (Fig. 6B). HA protein was observed along the apical membrane (Fig. 6B), and the NP and M1 proteins were detected near the apical membrane containing HA (Fig. 6C). The distributions of these major viral proteins were quite similar between the BM2stop mutant and rg-B/Y wt viruses.
Viral components in virions of BM2 knockout virus.
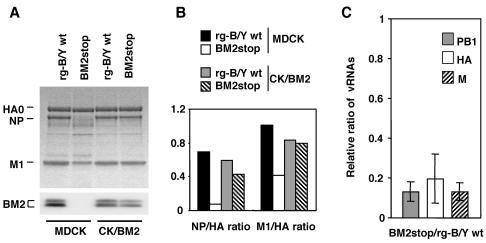
It is of interest to know why the BM2stop mutant virus did not grow in the absence of BM2 and how this mutant virus regained its ability to grow by supplementation of BM2. To clarify these issues, the BM2stop mutant and rg-B/Y wt viruses were infected into MDCK and CK/BM2 cells at a multiplicity of infection (MOI) of 5 PFU per cell, and progeny viruses released into the culture supernatant were purified and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 7). In the virion of the mutant virus produced by MDCK cells, the BM2 was not detected at all and NP was not clearly detected in the virions. Moreover, the content of M1 normalized with that of HA was found to be less than half of that in rg-B/Y wt virus (Fig. 7B). In contrast, in the virion of mutant viruses produced by CK/BM2 cells, NP and M1 were recovered at about 80 and 90%, respectively, of the levels in rg-B/Y wt virus (Fig. 7A and B). These results indicate that the incorporation of M1 and NP into virions was highly affected in the absence of BM2. Interestingly, comparable amounts of BM2 supplied by CK/BM2 cells were also incorporated into the virions.
FIG. 7.
Viral proteins and RNA segments in BM2stop mutant virus particles. (A) Protein composition of BM2stop mutant and rg-B/Y wt virions. The purified virions that were produced by MDCK and CK/BM2 cells were analyzed by Coomassie brilliant blue staining (upper panel) and Western blotting with anti-BM2 antibody (lower panel). (B) Relative amounts of viral proteins. Viral proteins were quantified by using an ATTO CS analyzer, and the relative staining intensity of each protein was normalized to that of HA (set at 1.00) for each virus. (C) Relative ratios of PB1, HA, and M genes between BM2stop and rg-B/Y wt virions. RNA was extracted from the virions grown in MDCK cells and reverse transcribed, and the resultant cDNA was quantitated by real-time PCR with primers directed to the PB1, HA, and M genes (see Materials and Methods). The mutant virions analyzed had amounts of HA proteins equal to those of rgB/Y wt. All samples were run in duplicate and repeated three times. Error bars represent the standard errors of the means.
Since NP, which constitutes the backbone of the vRNP, was reduced in the mutant virions, we speculated that BM2 is required for the packaging of vRNP. Further studies were therefore conducted to determine whether incorporation of vRNA into virions was affected by the absence of BM2 expression. RNA was extracted from the purified BM2stop mutant and rg-B/Y wt virions grown in MDCK cells. The mutant virions used in this experiment had an amount of HA proteins equal to that of rgB/Y wt virions. After extraction, vRNA was reverse transcribed and the resultant cDNA was analyzed by real-time PCR with primers directed to the PB1, HA, and M genes (see Materials and Methods). Copy numbers of the vRNA detected in the mutant virions were approximately eightfold lower for the PB1 and M genes and about fivefold lower for than HA gene than were those of the rg-B/Y wt virions (Fig. 7C). This result indicated that the decrease in NP and M1 is the result of inhibited incorporation of the vRNP complex into the virion. Taken together, the data suggest that BM2 is responsible for the incorporation of the vRNP complex into virion.
Truncated BM2 affects incorporation of vRNP into virions.
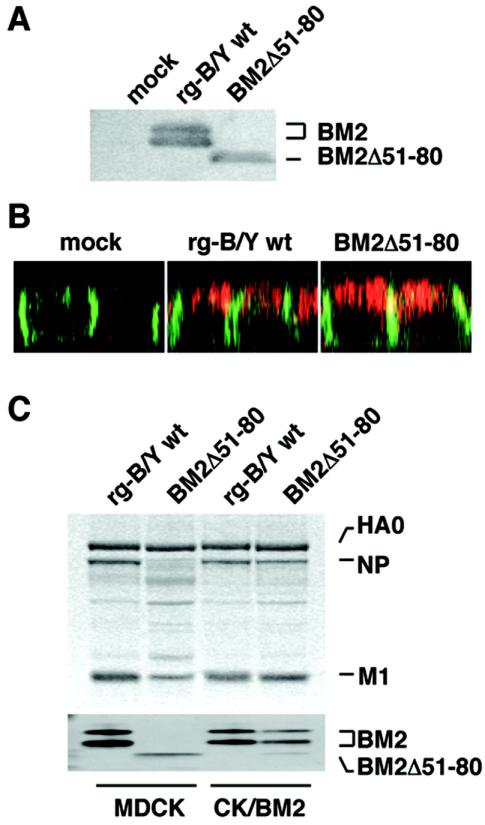
It was previously shown that the truncated BM2 (deletion from amino acids [aa] 51 to 80) was transported to the plasma membrane as normally seen in wt BM2 (32). The BM2Δ51-80 mutant virus, however, was not recovered by the reverse genetics system (Table 1) but was recovered when wt BM2 was supplemented from CK/BM2 cells (data not shown). The recovered mutant virus synthesized a truncated BM2 in infected MDCK cells (Fig. 8A), and this product was found at the apical membrane, similar to that of wt BM2, in an XZ section by IFA (Fig. 8B).
FIG. 8.
Characterizations of viral proteins of BM2Δ51-80 mutant viruses in infected cells and purified virions. (A) BM2 proteins in MDCK cells were analyzed by Western blotting with anti-BM2 antibody. (B) The localization of BM2 proteins at the apical membrane of MDCK cells was examined by IFA with anti-BM2 antibody (BM2 is shown in red) together with anti-β-catenin antibody (β-catenin is shown in green). (C) The protein composition of the mutant and rg-B/Y wt virions was analyzed as described for Fig. 7.
The components of the purified virion of BM2Δ51-80 mutant virus were also examined in parallel with that of rg-B/Y wt virus (Fig. 8C). In the virions produced by MDCK cells, dramatic reductions in NP and M1 were observed, although the truncated BM2 was incorporated into the virions. In contrast, in the virions produced by CK/BM2 cells, the amounts of NP and M1 were approximately 70 and 90%, respectively, of the levels seen in rg-B/Y wt virus. These observations were consistent with the findings for BM2stop mutant virus, indicating that the loss of intrinsic BM2 expression and the loss of BM2 itself resulted in the suppression of vRNP incorporation into virions. Interestingly, the BM2Δ51-80 mutant virus grown in CK/BM2 cells contained two species of BM2, the wt BM2 supplied from CK/BM2 host cells and the truncated BM2, in different amounts. The levels of incorporation of these proteins were reflected by the levels of their syntheses in infected cells.
Membrane association of M1 is greatly suppressed in the absence of BM2.
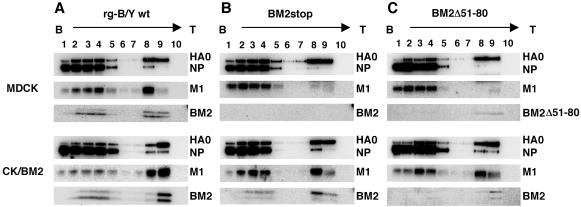
During the assembly process, viral components are transported to the budding site of the plasma membrane and interact directly or indirectly with the membrane (2). To clarify why incorporation of vRNP into virions was greatly affected in the absence of BM2, the membrane association of the viral proteins was examined by sucrose flotation centrifugation analysis. Figure 9 shows the fractions of infected cell lysates collected at 11 hpi followed by Western blotting with anti-B/Yamagata and anti-BM2 antibodies. Fractions 8 and 9 were found to mainly contain cellular membranes in a previous study (32). In MDCK cells, at least half of the total amount of HA and M1 of rg-B/Y wt virus was detected in the membrane fractions and part of NP was also found in fraction 8 (Fig. 9A, top panel). In contrast, the majority of the M1 in the BM2stop and BM2Δ51-80 mutant viruses was found in cytosolic fractions 1 to 4 and only a trace amount of M1 was detected in the membrane fractions, although the membrane-associated HA did not greatly differ from that in rg-B/Y wt virus (Fig. 9B and C, top panels). The substantial reduction in membrane-associated M1 may correspond to the decrease in NP, probably vRNP, in the membrane fractions. On the other hand, with trans-supplementation of BM2 from CK/BM2 host cells, no suppression of the membrane association of M1 was observed (Fig. 9B and C, bottom panels). Because the distribution of M1 from BM2stop mutant viruses at the apical membrane of MDCK cells did not differ from that of rg-B/Y wt virus (Fig. 6C), these results indicate that the membrane association of M1 was highly affected in the absence of BM2 on the apical membrane, resulting in the failure of packaging of the vRNP complex into the virions.
FIG. 9.
Membrane association of viral proteins in virus-infected cells. MDCK and CK/BM2 cells were infected with rg-B/Y wt (A), BM2stop (B), and BM2Δ51-80 (C) viruses. At 11 hpi, infected cells were lysed and postnuclear fractions were subjected to equilibrium centrifugation as described in Materials and Methods. Viral proteins in all fractions collected from the top (T) to the bottom (B) were analyzed by Western blotting with anti-B/Yamagata and anti-BM2 antibodies.
DISCUSSION
BM2 is an integral membrane protein that is transported to the plasma membrane (28, 32) and is incorporated into virions as a minor component (27). Electrophysiological studies of BM2 in Xenopus oocytes and in mammalian cells demonstrated that BM2 has proton (H+) ion channel activity as a functional counterpart of the influenza A virus M2 protein (24). To further explore the role of BM2 in the life cycle of influenza B virus, we generated knockout and truncated mutant viruses of BM2 by using a newly developed reverse genetics system for influenza B virus.
The BM2ΔATG mutant virus, in which the initiation codon for BM2 translation (771AUG) was replaced with 771ACC, was recovered at levels similar to those of the wt virus from plasmid-transfected cells (Table 1). This mutant virus grew more slowly than wt virus in the multiple-step growth cycle but produced progeny viruses with a maximum yield similar to that of wt virus (Fig. 2). Although the BM2 ORF of the mutant virus did not contain an initiation codon, BM2 was synthesized at low levels in infected cells and was detected in the virions (Fig. 2). BM2 and influenza A virus M2 have been shown to have similar ion channel activities (24). The ion channel of influenza A virus M2 is thought to be important for dissociation of M1 from RNP by weakening the protein-protein interactions by acidification from pH 7.4 to 5.8 during uncoating (5, 6, 22, 39, 41), resulting in the subsequent transport of vRNP into the nucleus, where replication and transcription of the virus genome occur (20). If BM2 ion channel activity is crucial, it is possible that inefficient uncoating as a result of reduced expression of the ion channel protein BM2 is one of the reasons for the slow growth of the BM2ΔATG mutant virus. However, the final virus yield of the mutant virus was similar to that of the wt virus, indicating that a minimum amount of BM2 can support full growth of influenza B virus.
The question of how BM2 is synthesized without the AUG initiation codon in the ORF thus arises. The synthesized BM2 had similar molecular mass, 12 kDa (15, 27), to wt BM2. In the +2 ORF of RNA segment 7, in which BM2 is translated (15), there are three AUG triplets, 831AUG, 1002AUG, and 1062AUG, which are potential initiation codons downstream of the 771AUG BM2 initiation codon, but there are none upstream. If translation occurred from any of these triplets, the products would lack the transmembrane domain (TMD), which is translated from 771AUG to 837TGG, like the BM2Δ2-23 mutant (32). The mutant lacking the TMD in the BM2 molecule was not recovered at all, as shown in Table 1, because the BM2 molecule without TMD is not properly transported to the budding site on the plasma membrane (32). Moreover, the putative molecular masses of the products would be 10, 3.6, or 1.4 kDa. These products are smaller than wt BM2, and if such small products are produced, they could be distinguished from wt BM2, indicating that these triplets are not used for BM2 synthesis in the BM2ΔATG mutant virus. Moreover, BM2 of the mutant virus reacted with an anti-BM2 antibody against wt BM2 protein and functioned as wt BM2 to produce infectious viruses, thus suggesting that the amino acid alignment between the BM2ΔATG mutant and wt viruses is the same. It is therefore most likely that the BM2 of the BM2ΔATG mutant virus is translated from the 771ACC replacement codon. Similar unusual translation initiated from GUG or UUG triplets is observed with Escherichia coli, although their translational efficiency is considerably lower than that initiated from the AUG triplet (19).
By inserting three consecutive stop codons downstream of the 771ACC replacement codon in BM2ΔATG plasmid DNA, a knockout virus of BM2 was successfully generated in a BM2 trans-supplementation system. This BM2 knockout virus permitted production of infectious viruses only when wt BM2 was supplemented by host cells expressing BM2 (Fig. 5). Consequently, it was apparent that BM2 is a necessary component for the production of infectious progeny viruses. Mould et al. (24) recently indicated that overexpression of BM2 causes morphological changes in the Golgi apparatus and a delay in exocytic transport of HA when BM2 and HA of influenza A/FPV (Rostock strain) virus were coexpressed in HeLa cells. However, we did not observe this disadvantageous virus growth because the growth of wt virus in the CK/BM2 cell line, in which BM2 was expressed by host cells in addition to the expression by virus itself, did not differ from that in MDCK cells (Fig. 5). Presumably, the expression level of BM2 does not influence the normal growth of influenza B virus in tissue culture.
Previously, Watanabe et al. (33) suggested that the ion channel activity of influenza A virus is dispensable for growth in tissue culture, whereas Takeda et al. (30) subsequently argued their findings by analysis of amantadine sensitivity and concluded that ion channel activity contributes to the efficient replication of virus. Although the ion channel activity of BM2 may be important for the growth of influenza B virus, our study of BM2 knockout virus suggested another function of BM2 in the life cycle of influenza B virus. In the absence of BM2 expression, the synthesis and cytoplasmic transport of major viral proteins, HA, NP, and M1, to the virion budding site occurred normally and were indistinguishable between the BM2 knockout and wt viruses (Fig. 6). The most striking difference was found in the content of the vRNP complex, NP and M1, in virions when comparing viruses produced in the absence and presence of BM2 (Fig. 7A). These differences were also found in the case of loss of intrinsic BM2 expression by internal deletion of the molecule, even when the truncated BM2 was synthesized and integrated into the plasma membrane, as occurs in the wt virus (Fig. 8C). Therefore, it is most likely that BM2 plays a role in the incorporation of the vRNP complex into the virion during the assembly process. This interpretation does not conflict with the observation that suppressed synthesis of wt BM2 correlates to a decrease in the content of vRNP, as calculated by the NP/HA ratio in the BM2ΔATG mutant virion (Fig. 2).
In influenza A viruses, M1 has lipid-binding properties (11, 29). The cytoplasmic tails of HA and NA glycoproteins on the plasma membrane are known to stimulate the membrane association of M1 to form a shell lining the inner surface of the viral envelope (7, 10). Unlike influenza A virus, the assembly mechanism of virus components at the virion budding site on the plasma membrane are not well characterized for influenza B virus. In the present study, we confirmed by XZ section observation of infected cells by confocal microscopy that influenza B virus M1 clearly underlined the apical membrane containing HA (Fig. 6). If the cytoplasmic tails of the envelope proteins HA, NA, and NB mediate the membrane association of M1, as in the influenza A virus, their interactions would not be so close. Our results with BM2 knockout virus indicated that the membrane association of influenza B virus M1 was highly influenced by the presence and the absence of BM2 on the membrane. In fact, flotation gradient centrifugation analysis revealed that the membrane association of M1 was greatly affected by the absence of BM2, whereas this decreased membrane association of M1 was restored when wt BM2 was supplemented in trans by host cells expressing BM2 (Fig. 9). Based on these results, it is likely that BM2 stimulates the binding of M1, presumably through an M1-BM2 interaction, to the apical membranes. This M1-BM2 interaction was suggested by our previous experiments in which BM2 was coprecipitated with M1 when detergent-disrupted virion was immunoprecipitated with an anti-M1 antibody, and in experiments with virion fractionation, BM2 always coexisted with M1 in the same fractions but was not present with HA and vRNP without M1 (27). Moreover, the longest cytoplasmic tail of BM2 may contribute to the capture of M1 at the membrane site because the cytoplasmic tails of BM2, HA, NA, and NB are deduced to be 84 to 86 aa residues (28, 32), 10 aa residues (8), 12 aa residues (8), and 60 aa residues (35), respectively. This interpretation is consistent with the result that the shortened cytoplasmic tail of BM2Δ51-80 mutant virus, which possesses a 30-aa deletion in the BM2 molecule, resulted in a failure to stimulate the membrane association of M1, similar to the BM2 knockout virus after flotation centrifugation (Fig. 9C). Taken together, these findings suggest that BM2 interacts with M1 at the plasma membrane and that this BM2-M1 interaction may be necessary for the tight association of M1 to the plasma membrane.
In the case of influenza A virus, M1 interacts with vRNP through direct binding with vRNA and interaction with NP (31, 36), and this complex formation is thought to be necessary to capture the vRNP complex at the virion budding site during the assembly process. Similarly, in the case of influenza B virus, vRNP was also suggested to form a complex with M1 by our virion fractionation experiments (16). This complex formation would also be necessary to capture vRNP at the virion budding site like influenza A virus. Consequently, the significant decrease in membrane binding of M1 in the absence of BM2 may result in reduced incorporation of the M1-vRNP complex into virions.
In the particles of the BM2stop mutant and BM2Δ51-80 mutant viruses, however, M1 was still detectable, although the content of M1 was greatly decreased together with the lack of vRNP incorporation into virion (Fig. 7 and 8). The results may imply that there are two forms of M1 in the virion, i.e., M1 with vRNP and M1 without vRNP. The lack of vRNP with the decrease of M1 in the virion in the absence of BM2 suggests that BM2 plays an important role in the tight membrane association and subsequent incorporation of the M1-vRNP complex during the virion assembly process. The precise function of BM2 on vRNP packaging into virions should be clarified to elucidate the mechanism of influenza B virus assembly.
Acknowledgments
We thank Y. Kawaoka (University of Wisconsin—Madison, Madison) and G. Hobom (Methesys GmbH, Cologne, Germany) for kindly providing the PolI vector and A. García-Sastre (Mount Sinai School of Medicine, New York, N.Y.) and N. Nakagawa (Institute of Public Health of Kobe City) for providing plasmid pCB7 and anti-B/HA mouse monoclonal antibody, respectively. We also thank A. Kato (National Institute of Infectious Diseases, Tokyo, Japan) for helpful advice with the real-time PCR experiment.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology and by a grant from the Ministry of Health, Labor, and Welfare.
REFERENCES
- 1.Ali, A., R. T. Avalos, E. Ponimaskin, and D. P. Nayak. 2000. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J. Virol. 74:8709-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barman, S., A. Ali, E. K. Hui, L. Adhikary, and D. P. Nayak. 2001. Transport of viral proteins to the apical membranes and interaction of matrix protein with glycoproteins in the assembly of influenza viruses. Virus Res. 77:61-69. [DOI] [PubMed] [Google Scholar]
- 3.Brassard, D. L., G. P. Leser, and R. A. Lamb. 1996. Influenza B virus NB glycoprotein is a component of the virion. Virology 220:350-360. [DOI] [PubMed] [Google Scholar]
- 4.Briedis, D. J., R. A. Lamb, and P. W. Choppin. 1982. Sequence of RNA segment 7 of the influenza B virus genome: partial amino acid homology between the membrane proteins (M1) of influenza A and B viruses and conservation of a second open reading frame. Virology 116:581-588. [DOI] [PubMed] [Google Scholar]
- 5.Bui, M., G. Whittaker, and A. Helenius. 1996. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J. Virol. 70:8391-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukrinskaya, A. G., N. K. Vorkunova, G. V. Kornilayeva, R. A. Narmanbetova, and G. K. Vorkunova. 1982. Influenza virus uncoating in infected cells and effect of rimantadine. J. Gen. Virol. 60:49-59. [DOI] [PubMed] [Google Scholar]
- 7.Enami, M., and K. Enami. 1996. Influenza virus hemagglutinin and neuraminidase glycoproteins stimulate the membrane association of the matrix protein. J. Virol. 70:6653-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flandorfer, A., A. García-Sastre, C. F. Basler, and P. Palese. 2003. Chimeric influenza A viruses with a functional influenza B virus neuraminidase or hemagglutinin. J. Virol. 77:9116-9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. García-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez-Puertas, P., C. Albo, E. Perez-Pastrana, A. Vivo, and A. Portela. 2000. Influenza virus matrix protein is the major driving force in virus budding. J. Virol. 74:11538-11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregoriades, A., and B. Frangione. 1981. Insertion of influenza M protein into the viral lipid bilayer and localization of site of insertion. J. Virol. 40:323-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiebert, S. W., M. A. Williams, and R. A. Lamb. 1986. Nucleotide sequence of RNA segment 7 of influenza B/Singapore/222/79: maintenance of a second large open reading frame. Virology 155:747-751. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann, E., K. Mahmood, C. F. Yang, R. G. Webster, H. B. Greenberg, and G. Kemble. 2002. Rescue of influenza B virus from eight plasmids. Proc. Natl. Acad. Sci. USA 99:11411-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann, E., G. Neumann, Y. Kawaoka, G. Hobom, and R. G. Webster. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 97:6108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horvath, C. M., M. A. Williams, and R. A. Lamb. 1990. Eukaryotic coupled translation of tandem cistrons: identification of the influenza B virus BM2 polypeptide. EMBO J. 9:2639-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imai, M., S. Watanabe, and T. Odagiri. 2003. Influenza B virus NS2, a nuclear export protein, directly associates with the viral ribonucleoprotein complex. Arch. Virol. 148:1873-1884. [DOI] [PubMed] [Google Scholar]
- 17.Jackson, D., A. Cadman, T. Zurcher, and W. S. Barclay. 2002. A reverse genetics approach for recovery of recombinant influenza B viruses entirely from cDNA. J. Virol. 76:11744-11747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin, H., G. P. Leser, J. Zhang, and R. A. Lamb. 1997. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 16:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozak, M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187-208. [DOI] [PubMed] [Google Scholar]
- 20.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1531. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 21.Lamb, R. A., S. L. Zebedee, and C. D. Richardson. 1985. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 40:627-633. [DOI] [PubMed] [Google Scholar]
- 22.Martin, K., and A. Helenius. 1991. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67:117-130. [DOI] [PubMed] [Google Scholar]
- 23.Mora, R., E. Rodriguez-Boulan, P. Palese, and A. García-Sastre. 2002. Apical budding of a recombinant influenza A virus expressing a hemagglutinin protein with a basolateral localization signal. J. Virol. 76:3544-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mould, J. A., R. G. Paterson, M. Takeda, Y. Ohigashi, P. Venkataraman, R. A. Lamb, and L. H. Pinto. 2003. Influenza B virus BM2 protein has ion channel activity that conducts protons across membranes. Dev. Cell 5:175-184. [DOI] [PubMed] [Google Scholar]
- 25.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 27.Odagiri, T., J. Hong, and Y. Ohara. 1999. The BM2 protein of influenza B virus is synthesized in the late phase of infection and incorporated into virions as a subviral component. J. Gen. Virol. 80:2573-2581. [DOI] [PubMed] [Google Scholar]
- 28.Paterson, R. G., M. Takeda, Y. Ohigashi, L. H. Pinto, and R. A. Lamb. 2003. Influenza B virus BM2 protein is an oligomeric integral membrane protein expressed at the cell surface. Virology 306:7-17. [DOI] [PubMed] [Google Scholar]
- 29.Ruigrok, R. W., A. Barge, P. Durrer, J. Brunner, K. Ma, and G. R. Whittaker. 2000. Membrane interaction of influenza virus M1 protein. Virology 267:289-298. [DOI] [PubMed] [Google Scholar]
- 30.Takeda, M., A. Pekosz, K. Shuck, L. H. Pinto, and R. A. Lamb. 2002. Influenza a virus M2 ion channel activity is essential for efficient replication in tissue culture. J. Virol. 76:1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe, K., H. Handa, K. Mizumoto, and K. Nagata. 1996. Mechanism for inhibition of influenza virus RNA polymerase activity by matrix protein. J. Virol. 70:241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe, S., M. Imai, Y. Ohara, and T. Odagiri. 2003. Influenza B virus BM2 protein is transported through the trans-Golgi network as an integral membrane protein. J. Virol. 77:10630-10637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe, T., S. Watanabe, H. Ito, H. Kida, and Y. Kawaoka. 2001. Influenza A virus can undergo multiple cycles of replication without M2 ion channel activity. J. Virol. 75:5656-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whittaker, G., M. Bui, and A. Helenius. 1996. Nuclear trafficking of influenza virus ribonucleoproteins in heterokaryons. J. Virol. 70:2743-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams, M. A., and R. A. Lamb. 1986. Determination of the orientation of an integral membrane protein and sites of glycosylation by oligonucleotide-directed mutagenesis: influenza B virus NB glycoprotein lacks a cleavable signal sequence and has an extracellular NH2-terminal region. Mol. Cell. Biol. 6:4317-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye, Z., T. Liu, D. P. Offringa, J. McInnis, and R. A. Levandowski. 1999. Association of influenza virus matrix protein with ribonucleoproteins. J. Virol. 73:7467-7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zebedee, S. L., and R. A. Lamb. 1988. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J. Virol. 62:2762-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zebedee, S. L., C. D. Richardson, and R. A. Lamb. 1985. Characterization of the influenza virus M2 integral membrane protein and expression at the infected-cell surface from cloned cDNA. J. Virol. 56:502-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, J., and R. A. Lamb. 1996. Characterization of the membrane association of the influenza virus matrix protein in living cells. Virology 225:255-266. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhirnov, O. P. 1992. Isolation of matrix protein M1 from influenza viruses by acid-dependent extraction with nonionic detergent. Virology 186:324-330. [DOI] [PubMed] [Google Scholar]