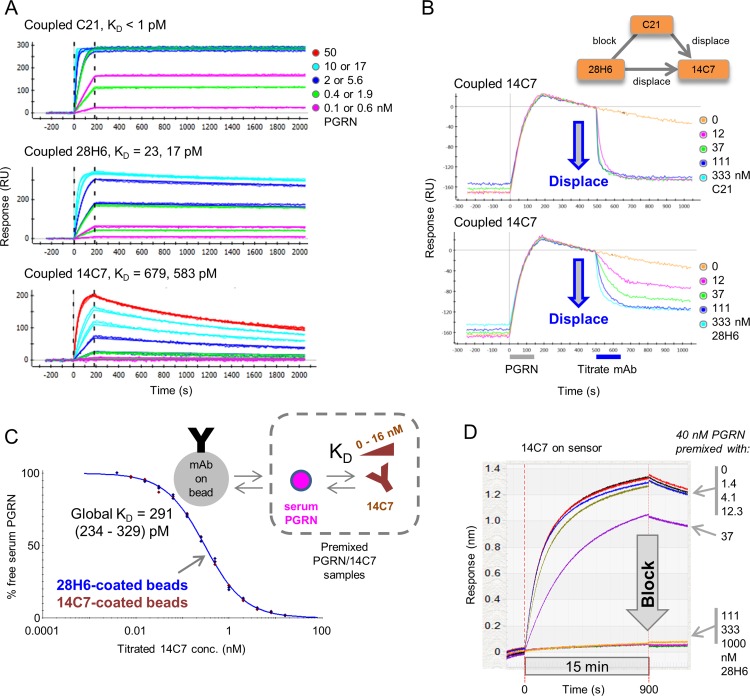
Fig 5. Kinetic and epitope binning characterization of anti-PGRN mAbs that displace one another.
(A) One-shot kinetic analysis of PGRN binding to coupled mAbs C21, 28H6 (M27), and 14C7 (M4); see Table 1. (B) Examples of waterfall competition plots for mAb C21 and mAb 28H6 (M27) analytes showing their displacement of mAb 14C7 (M4) ligand (ProteOn data). Blocking relationships for these three mAbs are shown in the inset; C21 and 28H6 block one another and potently displace 14C7, whereas 14C7 blocks (barely displaces) C21 and 28H6. (C) KinExA analysis determining the apparent affinity of 14C7 for serum PGRN as detected using 14C7-coated or 28H6-coated beads. Curves from both bead types are overlaid and fit globally. (D) Premix competition of 14C7 and 28H6 using an Octet-Red384, showing that solution-based 28H6 at concentrations > 40 nM fully blocks binding of 40 nM to 14C7-coated sensors, consistent with an exact titration of binding sites.