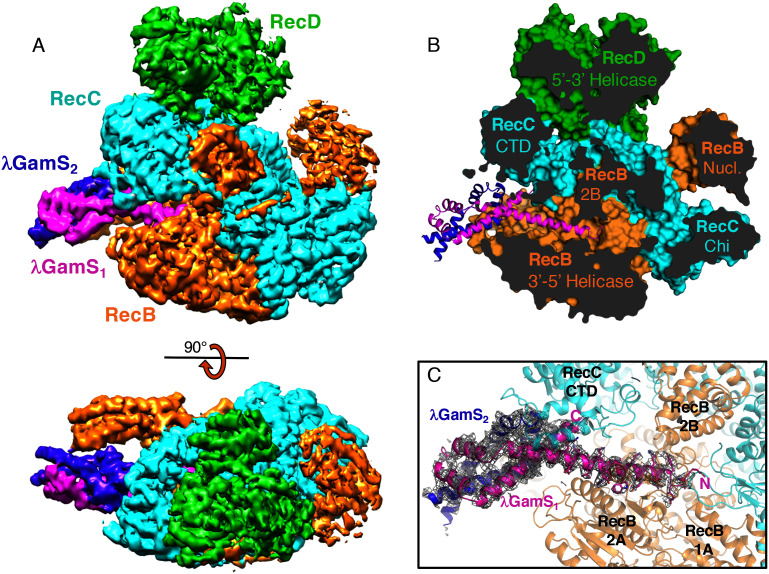
Figure 1. The overall structure of the RecBCD/Gam complex.
(A) Surface representation of the electron density with a ribbon representation of the RecBCD subunits and the GamS protein dimer. (B) Cut away of the molecular surface of the RecBCD part of the model with the GamS dimer overlaid showing how the protein enters and fills the channel normally occupied by the 3’ssDNA tail. (C) The same view with the electron density for the GamS dimer overlaid.
DOI: http://dx.doi.org/10.7554/eLife.22963.002