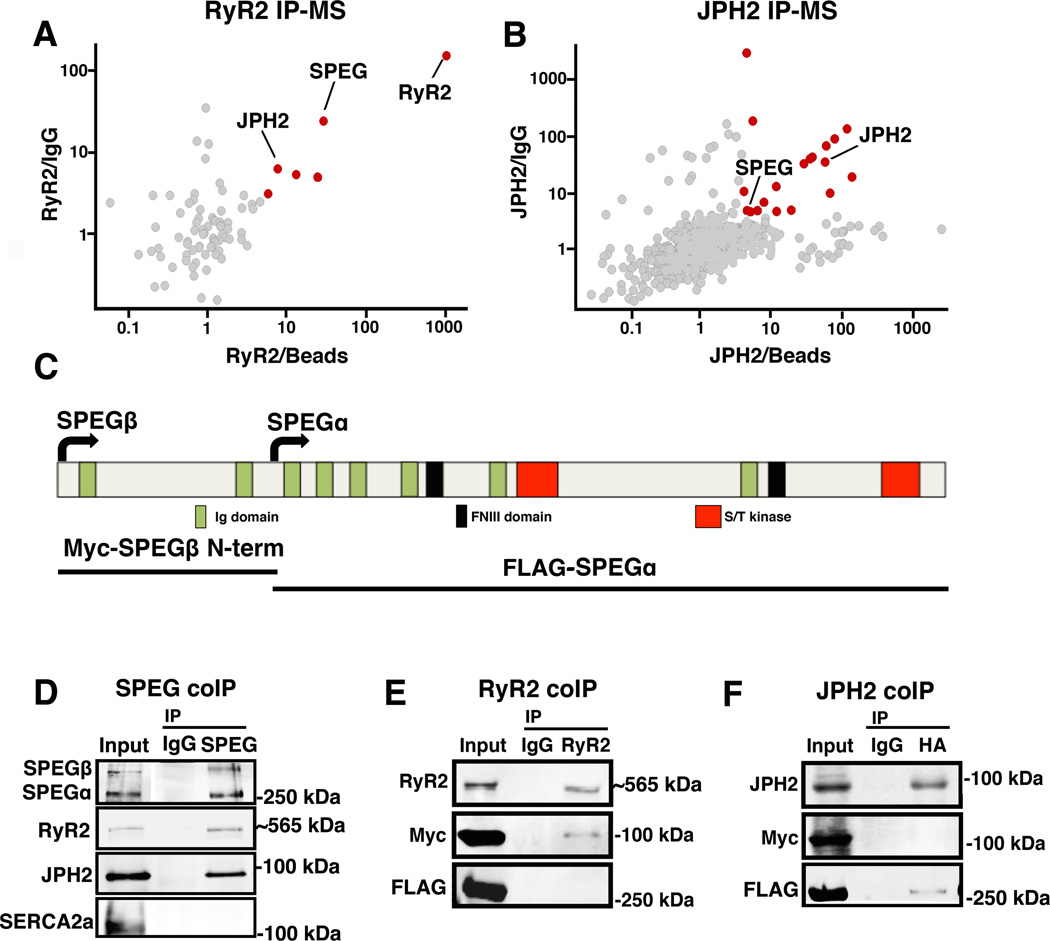
Figure 1. SPEG directly binds to RyR2 and JPH2 within JMCs.
(A–B) Unbiased proteomics analyses of putative RyR2 and JPH2 binding partners based on adult mouse heart co-immunoprecipitation of RyR2 and JPH2, respectively, coupled to mass spectrometry (IP-MS). Specific binding partners (red dots) were defined based on cut-off ratios of 3 and 4, respectively, of the MS intensity of each protein that came down with RyR2 or JPH2 IP vs. control IgG IP (y-axis) and RyR2 or JPH2 IP vs. beads only IP (x-axis). (C) Schematic overview of SPEG protein structure with key functional domains marked, and SPEG fragments used for binding-site analysis. (D) Western blots of reciprocal SPEG co-IP experiment from mouse heart showing that both RyR2 and JPH2 but not SERCA2a pulled down with SPEG. (E) RyR2, Myc-tagged SPEGβ N-terminal peptide, and FLAG-tagged SPEGα were co-expressed in HEK293 cells. Only SPEGβ N-terminal peptide was shown to bind the RyR2 pull-down. (F) JPH2 was co-expressed with Myc-tagged SPEGβ N-terminal peptide and FLAG-tagged SPEGα; only FLAG-tagged SPEGα was shown to bind the JPH2 pull-down.