Figure 1. Directed evolution of cas9 generates mutants with increased specificity for NAG targets.
See also Figure S1.
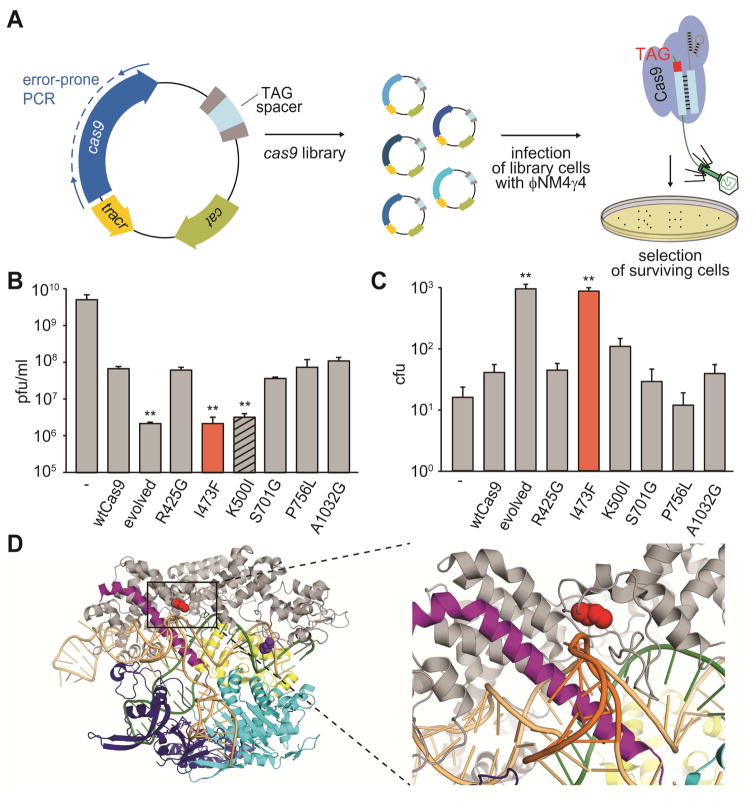
(A) Schematic diagram of the directed evolution assay. S. pyogenes cas9 was mutagenized by error-prone PCR and library amplicons were cloned into a plasmid carrying a spacer matching a TAG-adjacent target sequence on the ϕNM4γ4 phage. Library cells were infected with lytic phage to screen for mutants displaying improved NAG cleaving efficiency.
(B) Phage propagation was measured as the number of plaque forming units (pfu) per ml of stock, on cells targeting the NAG-adjacent protospacer and harboring plasmids with different mutations on cas9: one of the “evolved” alleles or each of the six mutations present in this allele. Mutations with pfu values significantly different than wild type are highlighted (**, p-value < 0.05 compared to wtCas9).
(C) Colony forming units (cfu) obtained after phage infection of naïve cells (not programmed to target any viral sequence) harboring plasmids with different mutations in cas9. Mutations with cfu values significantly different than wild type are highlighted.
(D) Location of residues I473 and K500 on the Cas9:single-guide RNA ribonucleoprotein (PDB 4UN3). Red, I473; purple, K500; orange, sgRNA; green, target DNA (the GG PAM highlighted in red); grey, alpha-helical (REC) lobe; yellow, HNH domain; light blue, RuvC domain; blue, PAM-interacting CTD.