Fig. 3.
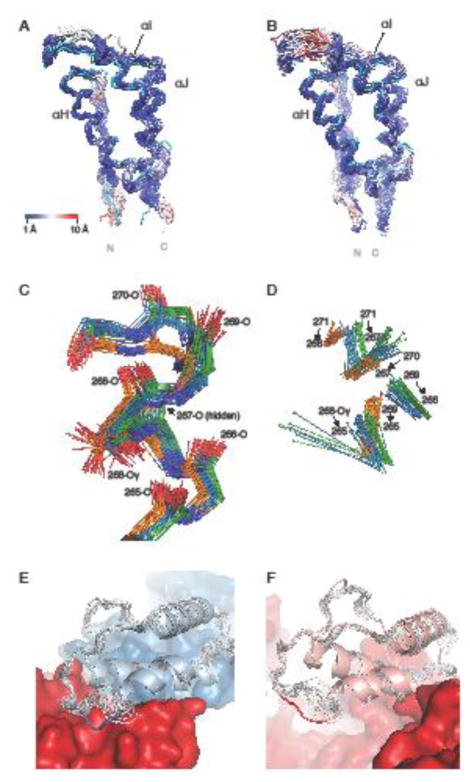
Comparison between the NMR structure of the isolated thumb domain and the thumb domain in the p51 and p66 chains in X-ray structures of the heterodimeric HIV-1 RT. a Backbone superposition of p51 thumb domain X-ray structures and the mean NMR structure (cyan), with atoms in the X-ray structures colored from blue to red with increasing RMSD values. b Equivalent superposition as in a, but for the p66 thumb domain X-ray structures. c Detailed view of residues 265-271, including the side chain of Ser268, for conformers in the NMR ensemble (orange carbons), p51 chains (blue carbons) and p66 chains (green carbons). d Hydrogen bonds between the H-atom of the donor (sphere) and the backbone oxygen acceptor in the NMR (orange), p51 (blue), and p66 (green) X-ray structures of the region shown in panel c. e Ribbon diagram of the p51 thumb domain (light blue) from a representative RT crystal structure (PDB Code 4IFY), with residues 285-287 depicted in blue. The remainder of p51 and p66 is shown in blue and red, respectively. The NMR ensemble (grey) differs from the X-ray structure around residue 287, where the p51 thumb subdomain contacts p66. f Ribbon diagram of the p66 thumb domain (pink) in a representative RT crystal structure (PDB Code 4IFY), highlighting the solvent exposed position of residues 285-287 (red coil). The remainder of the p66 chain is shown in red surface representation. In the NMR ensemble (grey), a similar local conformation is seen for residues 285-287.