Abstract
Proteolysis of autoantigens can alter normal MHC class II antigen processing and has been implicated in the induction of autoimmune diseases. Many autoantigens are substrates for the protease granzyme B (GrB), but the mechanistic significance of this association is unknown. Peptidylarginine deiminase 4 (PAD4) is a frequent target of autoantibodies in patients with rheumatoid arthritis (RA) and a substrate for GrB. RA is strongly associated with specific MHC class II alleles, and elevated levels of GrB and PAD4 are found in the joints of RA patients, suggesting that GrB may alter the presentation of PAD4 by RA-associated class II alleles. In this study, complementary proteomic and immunologic approaches were utilized to define the effects of GrB cleavage on the structure, processing, and immunogenicity of PAD4. Hydrogen–deuterium exchange and a cell-free MHC class II antigen processing system revealed that proteolysis of PAD4 by GrB induced discrete structural changes in PAD4 that promoted enhanced presentation of several immunogenic peptides capable of stimulating PAD4-specific CD4+ T cells from patients with RA. This work demonstrates the existence of PAD4-specific T cells in patients with RA and supports a mechanistic role for GrB in enhancing the presentation of autoantigenic CD4+ T cell epitopes.
Keywords: rheumatoid arthritis, peptidylarginine deiminase, shared epitope, autoimmunity, autoantigen, hydrogen–deuterium exchange, mass spectrometry, antigen processing, antigen presentation, epitope
Graphical abstract
INTRODUCTION
Modified self-proteins have the capacity to elicit autoimmune responses. The list of modifications linked to the development of autoimmune diseases is growing and includes nonsynonymous mutations in the setting of cancer,1,2 post-translational modifications,3,4 and proteolysis.5 Proteolysis of self-proteins has been linked to the development of autoimmune diseases, and fragments of autoantigens have been shown to be more immunogenic than their full-length counterparts.1,6,7 Many autoantigens across the spectrum of autoimmune diseases are susceptible to proteolysis by the serine protease granzyme B (GrB).8,9 Cytotoxic T lymphocytes (CTLs) deploy GrB to kill virally infected and transformed cells,10 and both infection and malignancy have been implicated as initiators of autoimmune diseases.2,3 Although GrB and caspases induce cell death by cleaving intracellular proteins after aspartic acid residues, GrB often yields novel protein fragments not observed during caspase-mediated apoptosis.8 A body of evidence suggests that GrB cleavage sites in autoantigens tend to colocalize with known T or B cell epitopes within unstructured protein loops,11 suggesting that this protease may have a role in the induction of autoimmunity, possibly through altering normal antigen processing and presentation.
The MHC locus is strikingly associated with the development of autoimmune diseases, and the vast majority of this risk is conferred by disease-specific MHC class II alleles at the HLA-DRB1 locus.12 MHC class II alleles are expressed by professional antigen presenting cells (APCs) that engulf extracellular antigens and serve to present processed peptides to CD4+ T cells. CD4+ T helper cells are therefore implicated as major drivers of autoimmune responses that secrete pro-inflammatory cytokines, provide help to CTLs to kill target cells, and stimulate B cells to make high-titer autoantibodies of the IgG subclass.13
The repertoire of peptides from a protein antigen that are ultimately presented in the groove of MHC class II molecules can be significantly affected by changes in antigen structure. Intact or partially unfolded protein antigens enter the MHC class II antigen processing pathway and can be protected from lysosomal cathepsin cleavage by binding to MHC class II molecules.14,15 Early proteolytic events, including those that occur prior to exposure to the antigen processing machinery,16–18 can affect these events and the nature of the peptides that are ultimately presented by MHC class II molecules. In addition, there is a hierarchy in how efficiently peptides from a protein are presented to T cells, such that not all possible peptides from a given antigen are encountered by CD4+ T cells during development.19 In the thymus, CD4+ T cells with high affinity for abundantly presented self-peptides (dominant peptides) are induced to die by apoptosis or rendered tolerant, whereas those recognizing poorly or never presented peptides (cryptic peptides) may survive and enter the circulation.19 T cells recognizing cryptic peptides generated under inflammatory conditions that modify antigen structure and processing have been hypothesized to contribute to the development of autoimmune diseases.20–22
Rheumatoid arthritis (RA) is a common systemic autoimmune disease in which GrB modification of antigens is suspected. RA is characterized by immune-mediated damage to synovial joints and is strongly associated with a specific group of MHC class II alleles called the “shared epitope” alleles (SE).3 The SE includes HLA-DR1 (DR1) and HLA-DR4 (DR4) and is associated with the development of autoantibodies to citrullinated proteins in patients with RA. Citrullinated proteins are generated through the post-translational modification of arginine residues to citrulline by peptidylarginine deiminase (PAD) enzymes.23 One of these enzymes, PAD4, is targeted by autoantibodies in 35% of patients with RA, and anti-PAD4 antibodies are associated with severe joint disease that progresses despite treatment.24,25 Despite the detection of high-titer anti-PAD4 IgG in these patients, PAD4-specific T cells have not yet been described. PAD4 is also cleaved by GrB at a single site, aspartic acid 388 (D388),25 and increased levels of GrB are observed in the joints and serum of RA patients with more erosive and progressive disease.26–29
Taken together, (i) the strong association of specific MHC class II molecules with the development of RA, (ii) the presence of anti-PAD4 antibodies and increased GrB levels in RA patients with severe disease, and (iii) susceptibility of PAD4 to proteolysis by GrB suggest that GrB might contribute to RA pathogenesis by exposing cryptic PAD4 epitopes capable of stimulating autoreactive T cells. To study this mechanism, a complementary set of proteomic and immunologic techniques were employed to define GrB-induced changes on PAD4 structure, processing, and immunogenicity. Hydrogen–deuterium exchange (HDX) coupled to tandem mass spectrometry (MS/MS) was utilized to assess dynamic changes in the PAD4 structure induced by GrB cleavage.30 In parallel, a minimalist cell-free MHC class II antigen processing system was utilized to define the effect of proteolysis by GrB on the processing and presentation of PAD4 peptides by the SE allele DR1.31 These approaches revealed that GrB-mediated cleavage of PAD4 induced discrete structural changes that enhanced the presentation of immunogenic PAD4 epitopes by DR1 and stimulated autoreactive T cells in the circulation of DR1/DR4-positive RA patients. This work suggests mechanism through which GrB contributes to the generation of cryptic epitopes in autoimmune disease.
MATERIALS AND METHODS
Study Approval and Statistical Analysis
PBMCs and autologous sera were obtained from patients with established RA, scleroderma, psoriatic arthritis, or healthy controls at the Johns Hopkins University under written informed consent under the auspices of the Johns Hopkins Institutional Review Board. RA patients were grouped according to the presence or absence of PAD4 peptide T cell responses, and clinical characteristics were compared. t tests were used for group-wise comparisons of normally distributed continuous variables; the Kruskal–Wallis test was used for group-wise comparisons of non-normally distributed variables; and two-sided Fisher’s Exact tests were used for group-wise comparisons of categorical variables. Analyses were performed using Graph-Pad Prism 6 and STATA software.
HLA-DR Genotyping
Intermediate resolution MHC class II genotyping was performed by the Johns Hopkins Immunogenetics Laboratory, and individuals with at least one copy of DRβ1*01 or DRβ1*04 were selected for subsequent studies.
Recombinant Proteins and Peptides
Recombinant N-terminal 6xHis tagged PAD4 (NP_036519) was expressed and purified from BL21(DE3) pLysS competent cells as previously described.25 The CEFT selected MHC class II peptide pool (PT-PA-CEFT-001-1) containing a mixture of CMV, EBV, influenza, and tetanus toxoid peptides was purchased from Axxora and served as a positive control for T cell activation. PAD4 peptides and a negative control peptide from RNA polymerase III (POLR3A, NP_008986) with known affinity for DR1 (IGTGLFKLLHKADRDPN)2 were synthesized by Elim Biopharmaceuticals to >90% purity.
Granzyme B Cleavage Assay
For T cell stimulation and MHC class II antigen processing assays, recombinant His-PAD4 was incubated at 37 °C for 1 h in the absence or presence of 80 nM recombinant human GrB in 1:10 diluted PBS. GrB alone and buffer only controls were also included. GrB cleavage was confirmed by separating proteins by SDS PAGE and subsequent Coomassie stain. For HDX assays, 20 µM His-PAD4 was diluted to 7.5 µM with reaction buffer (7.5 mM Tris, 150 mM NaCl, 0.4 mM EDTA, 4% glycerol, 4 mM β-mercaptoethanol) and incubated with 3.25 µM GrB for 2 h at 37 °C.
Hydrogen–Deuterium Exchange Analysis
Bottom-up HDX was performed using methods similar to previous descriptions.32 A comprehensive PAD4 peptide map (described in more detail in “Supporting Information (SI) methods”) was generated by combining peptides detected from intact and GrB-cleaved PAD4 and provided nearly complete coverage of PAD4, allowing HDX data to be obtained over the entire molecule. For HDX MS experiments, intact or cleaved PAD4 was HDX-labeled by incubating in D2O reaction buffer (7.5 mM Tris, 150 mM NaCl, 0.4 mM EDTA, 4% glycerol, 4 mM β-mercaptoethanol) for 10, 30, 60, 300, 900, or 3600 s at 20 °C in quadruplicate, quenched with 3 M urea/1% v/v trifluoroacetic acid, and digested at 1 °C prior to LCMS analysis. Peptides were separated by C8 trap and C18 column with 9.5 min HPLC (5 to 15% B over 0.1 min, linear increase to 50% B over 5.4 min, then to 98% B within 0.5 min; Eksigent) and analyzed by ESI high-resolution MS (m/z 300–2000) on an Exactive orbitrap mass spectrometer (Thermo Scientific) at 50 µL/min. The percent deuteration (%D) of each peptide was determined for each time point by normalizing measured deuterium content to the expected deuterium of 100% deuterated peptide and correcting for an estimated average deuterium recovery of 70% using the software HD Desktop.33 %D was plotted over HDX incubation time for each peptide of intact and GrB-cleaved PAD4 and analyzed by two-way ANOVA to identify peptides that underwent significant changes in deuteration following GrB cleavage. The %D difference was calculated by subtracting the aggregated %D values of intact PAD4 peptides from those of GrB-cleaved PAD4 over the on-exchange time from 10 to 3600 s.33
Cell-Free MHC Class II Antigen Processing System
Intact PAD4 or GrB-cleaved PAD4 were incubated with recombinant HLA-DR1 for 3 h at 37 °C in citrate buffer pH 5.0 as previously described.31 Briefly, Cathepsins B (bovine spleen, Sigma), H (human liver, Calbiochem), and S (human recombinant produced in E. coli, Calbiochem) were then added and incubated for an additional 2 h with 6 mM l-cysteine and 4 mM EDTA. The reaction was quenched by raising the pH to 7.5 and adding 10 mM iodoacetamide prior to immunoprecipitation of DR1/peptide complexes with anti-HLA-DR antibody (L243). Bound peptides were eluted with 1% TFA and filtered through a 10 kDa column to isolate peptides. Peptides were analyzed by liquid chromatography/tandem mass spectrometry (LC–MS/MS) using an LTQ Orbitrap Velos (Thermo Scientific) interfaced with a nanoAcquity UPLC system (Waters). Precursor and the fragment ions were analyzed at resolutions of 60,000 and 15,000, respectively, using the 371 m/z siloxane ion for a lock mass. Peptides were identified from Mascot (v2.5.1, Matrix Sciences) searches through the Proteome Discoverer (v1.4, Thermo Scientific) against the sample’s species proteins in RefSeq 2012 database with a 1% false discovery rate confidence threshold based on concatenated reverse database searches (see “SI Methods” for additional mass spectrometry methods). For analysis of the nested sets of peptides, the areas (ion counts × second) of each peptide in the nested set were calculated and summed. This total nested peak area was normalized to the total nested peak area of a DR1 peptide set to account for the amount of DR1 added to each sample.
PBMC Stimulation and Flow Cytometry
PBMCs from patients with RA, scleroderma, psoriatic arthritis, and healthy controls were isolated by Ficoll density centrifugation and plated at 1.5 × 106 cells per well in a flat-bottom 96 well plate in RPMI supplemented with 5% autologous serum, as previously described.2 In brief, anti-CD40 antibody was added to cultures to maintain cell-surface expression of CD154. For whole protein experiments, cells were stimulated with 15 µg/mL of intact PAD4, 15 µg/mL GrB-cleaved PAD4, GrB alone, or reaction buffer for 18 h at 37 °C. For peptide experiments, cells were stimulated with 2.5 µM PAD4 or control peptides for 18 h at 37 °C. Cells were harvested, washed in PBS, and stained with anti-CD3 BV510, anti-CD4 Pacific Blue, anti-CD8 APC-H7, anti-CD154 PE, and Invitrogen blue LIVE/DEAD stain prior to flow cytometric analysis. The % live CD3+, CD4+, CD8−, and CD154+ lymphocytes were determined using FCS express 5 flow research analysis software. The % CD4+CD154+ T cells observed over background activation to buffer alone (PAD4 protein studies) or the negative control POLR3A peptide (PAD4 peptide studies) was reported.
Protein Structure Images
Structures were made by overlaying the corresponding color codes for %D, %D difference, or PAD4 peptides identified by the cell-free system onto the calcium-free crystal structure of PAD4, protein data bank (PDB) ID 1WD834 using the PyMOL Molecular Graphics System, version 1.2r3pre (Schrödinger, LLC).
RESULTS
GrB Cleavage of PAD4 Enhanced Immunogenicity in a Subset of RA Patients
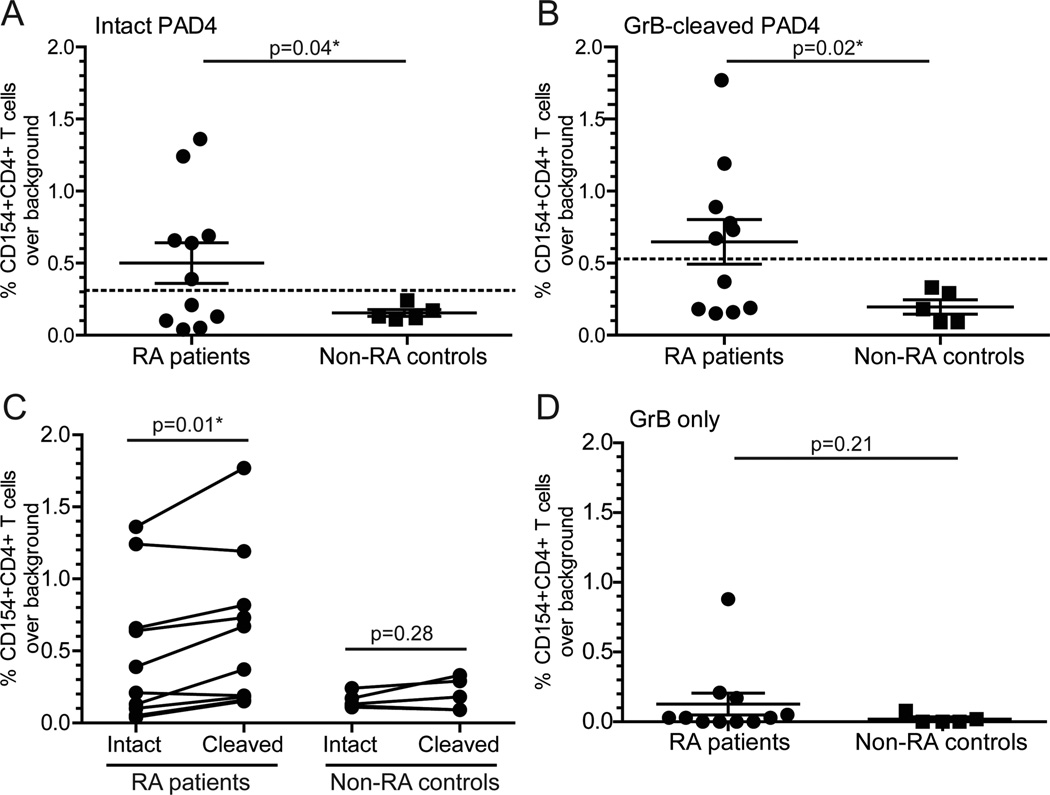
Circulation of anti-PAD4 IgG in a subset of patients with RA24,25 suggests the existence of PAD4-specific CD4+ T helper cells, although such cells have not been described to date. For determining if RA patients have PAD4-specific T cells, peripheral blood mononuclear cells (PBMCs) from RA patients (n = 11), healthy controls (n = 2), and scleroderma disease controls (n = 5) were stimulated with intact PAD4, and upregulation of the early activation marker CD154 was measured by flow cytometry. Using a cutoff of three standard deviations above the mean of the non-RA controls, PAD4-reactive T cells were detected in PBMCs from 6/11 (54%) patients with RA, and RA patients had a significantly higher percentage of CD4+CD154+ T cells (p = 0.03; Figure 1A) compared to non-RA controls following stimulation with intact PAD4.
Figure 1.
GrB-cleaved PAD4 is more immunogenic in a subset of patients with RA. PBMCs from patients with RA (n = 11) and non-RA controls (n = 2 healthy and n = 3 scleroderma controls) were stimulated with intact PAD4 (A), GrB-cleaved PAD4 (B), and GrB only (D) for 18 h. CD154 upregulation on CD4+ T cells was determined, and the % CD154+CD4+ T cells for each treatment condition minus background stimulation with buffer alone is shown on the y-axis. The threshold for a positive T cell response was set at three standard deviations above the mean of the non-RA controls for intact PAD4 (— at 0.31) or cleaved PAD4 (— at 0.53). (C) The change in % CD154+CD4+ T cells among individual RA patients (n = 10) and controls (n = 5) in response to intact vs cleaved PAD4 is shown. The RA patient with a background response to GrB alone was omitted because the independent response to GrB-cleaved PAD4 products could not be determined. Data were analyzed by an unpaired two-tailed t test with Welch’s correction (A–C) or a paired Student’s t test (D): * represents a significant p-value of <0.05.
In parallel, the effect of GrB cleavage of PAD4 on immunogenicity was explored by stimulating RA patient and non-RA control PBMCs with GrB-cleaved PAD4 or GrB alone. Following stimulation of PBMCs with GrB-cleaved PAD4, the same 6/11 (54.5%) RA patients upregulated CD154, and the mean percentage of activated CD154+CD4+ T cells was significantly increased compared to non-RA controls (p = 0.02; Figure 1B). However, RA patient T cells responded more robustly to GrB-cleaved PAD4 than to intact PAD4 (p = 0.01; Figure 1C). This trend was not observed with non-RA control cells (p = 0.28; Figure 1C), and there was no significant difference in response to GrB alone (p = 0.21; Figure 1D). This suggests that GrB-cleaved PAD4 is more immunogenic in a subset of patients with RA that have circulating PAD4-specific T cells, possibly due to alterations in antigen processing and exposure of cryptic PAD4 epitopes.
GrB Cleavage Induced Discrete Structural Changes in PAD4
To determine if proteolysis by GrB induces changes in the PAD4 protein structure that may alter MHC class II antigen processing, the effect of GrB cleavage on PAD4 protein structure was explored using HDX. HDX is a method for obtaining structural information on dynamic processes, including real-time conformational changes that are not amenable to study by X-ray crystallography.30,32 Structural information is obtained based on the rate at which amide hydrogens (–NH) in the protein backbone are exchanged for deuterium (a heavy isotope of hydrogen) atoms in a solution of heavy water (D2O). The rate of HDX is highly dependent on the local protein structure with flexible unstructured portions of a protein undergoing rapid rates of exchange and highly organized regions with extensive hydrogen bonding undergoing slower rates of exchange.35 This powerful method can identify regions of the PAD4 protein that become unstructured following proteolysis by GrB.
Following HDX, the PAD4 protein was digested and analyzed by MS to measure the incorporation of deuterium, indicated by a shift in one mass unit per hydrogen exchanged. The peptide profile generated for intact PAD4 demonstrated complete coverage of the protein (Figure S1), and consistent with the known dimeric organized protein structure,34 HDX-MS revealed that most regions of PAD4 exhibited low rates of deuterium exchange. Highly structured areas such as α-helices and β-sheets (Figure S2) were the most protected from HDX, whereas flexible loops, linker regions, and calcium binding sites had higher rates of HDX. The GrB cleavage site in PAD4 at D388, located within a surface-exposed disordered loop too unstructured to obtain crystallographic data,25,34 exhibited >90% HDX.
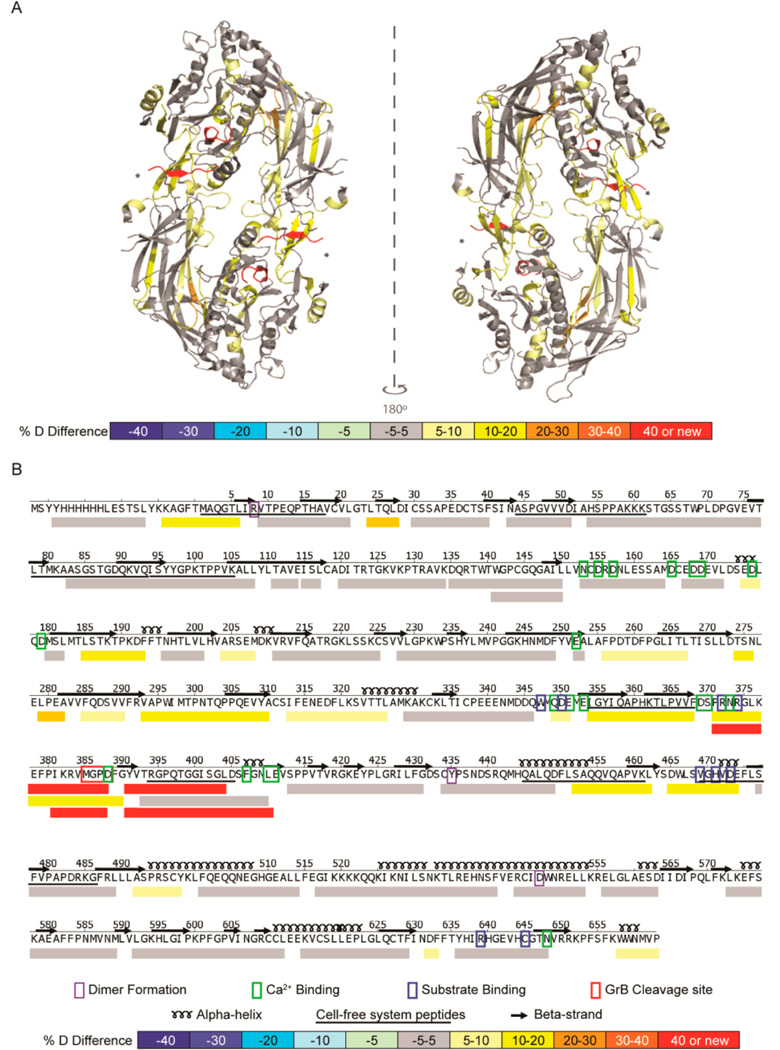
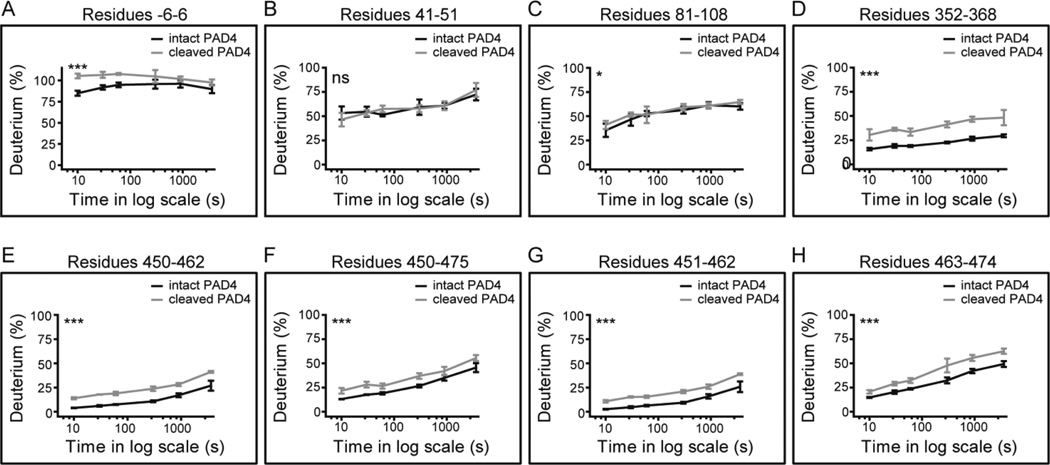
The HDX profile of GrB-cleaved PAD4 was compared with that of intact PAD4 to identify regions that became disordered following cleavage (Figure 2A). This confirmed that GrB cleaved PAD4 exclusively at D388 (Figure 2A). Novel peptides with HDX rates of >90% at F389–L410 and P372–D388 were detected at this site, corresponding to the newly created flexible N- and C-termini (Figure 2B). The most significant structural changes in PAD4 occurred within 150 amino acids of the GrB cleavage site, at the interface between the N- and C-terminal domains as well as in residues important for calcium ion and substrate binding (Figure 2B). Although specific regions of PAD4 became more disordered following proteolysis by GrB, the majority of the PAD4 structure remained unchanged. Of 50 representative PAD4 peptides that provide full protein sequence coverage, only 19 (38%) demonstrated statistically significant increases in structural dynamics following cleavage with GrB (p < 0.001; Figure 3 and Figure S3). HDX revealed that cleavage of PAD4 by GrB at D388 induced discrete structural changes in regions both adjacent to and remote from the GrB cleavage site. Importantly, GrB-induced changes in the PAD4 protein structure may alter downstream processing and lead to the presentation of cryptic PAD4 peptides.
Figure 2.
GrB cleavage of PAD4 induced structural changes adjacent to and remote from the cleavage site. (A) A differential HDX profile was generated indicating regions of the PAD4 protein that were exposed following proteolysis by GrB and was superimposed onto the calcium-free PAD4 dimeric crystal structure (PDB ID: 1WD8).34 The GrB cleavage site (*) demonstrates increased structural dynamics with the formation of new N- and C-termini (shown in red). (B) The linear differential HDX profile indicates the secondary structure elements (α-helices and β-strands), residues important for dimer formation (purple box), calcium binding (green box), and substrate binding (blue box), as well as the GrB cleavage site (red box). The %D difference was calculated by subtracting the %D observed for each peptide derived from intact PAD4 from that observed from peptides derived from GrB-cleaved PAD4 and is represented by a color-coded scale. Peptides identified by the cell-free MHC class II antigen processing system are underlined. HDX–MS was performed on intact and GrB-cleaved PAD4 in quadruplicate.
Figure 3.
GrB induced significant changes in the structural dynamics of distinct peptides located throughout the PAD4 protein. (A–H) HDX data of GrB-cleaved PAD4 (light gray line) and intact PAD4 (black line) are plotted as % incorporation of deuterium as a function of incubation time on a log scale in seconds (s). Data were analyzed by two-way ANOVA analysis: *** represents p < 0.001; * represents p ≤ 0.05, and ns represents a non-significant difference. The error bars represent the standard deviations from four independent HDX experiments. The individual PAD4 peptides shown are indicated by residue number and are 50 representative peptides identified in both the intact and cleaved PAD4 samples (additional data shown in Figure S3).
GrB Cleavage of PAD4 Enhanced Presentation of GrB-Exposed Peptides
Studies have shown that even minor changes to the three-dimensional structure of a protein antigen can alter its processing and presentation of epitopes.16–18 As such, flexible strands in a protein are more accessible to the antigen processing machinery and are susceptible to proteolysis by antigen processing enzymes or capture by MHC class II molecules.14–16,36 For assessing the effects that GrB-induced structural changes have on PAD4 antigen processing, a reductionist cell-free MHC class II antigen processing system, which has proven to be sufficient for processing of full-length antigens and identifying immunodominant epitopes, was utilized.18,31 This system is composed of only four purified components: an MHC class II molecule (in this case, the RA risk allele DR1) and three cathepsins (B, H, and S). Combined with high-resolution mass spectrometry for analysis of the peptides eluted from MHC class II molecules, the results provide clear answers about the processing and presentation of peptides from protein antigens.
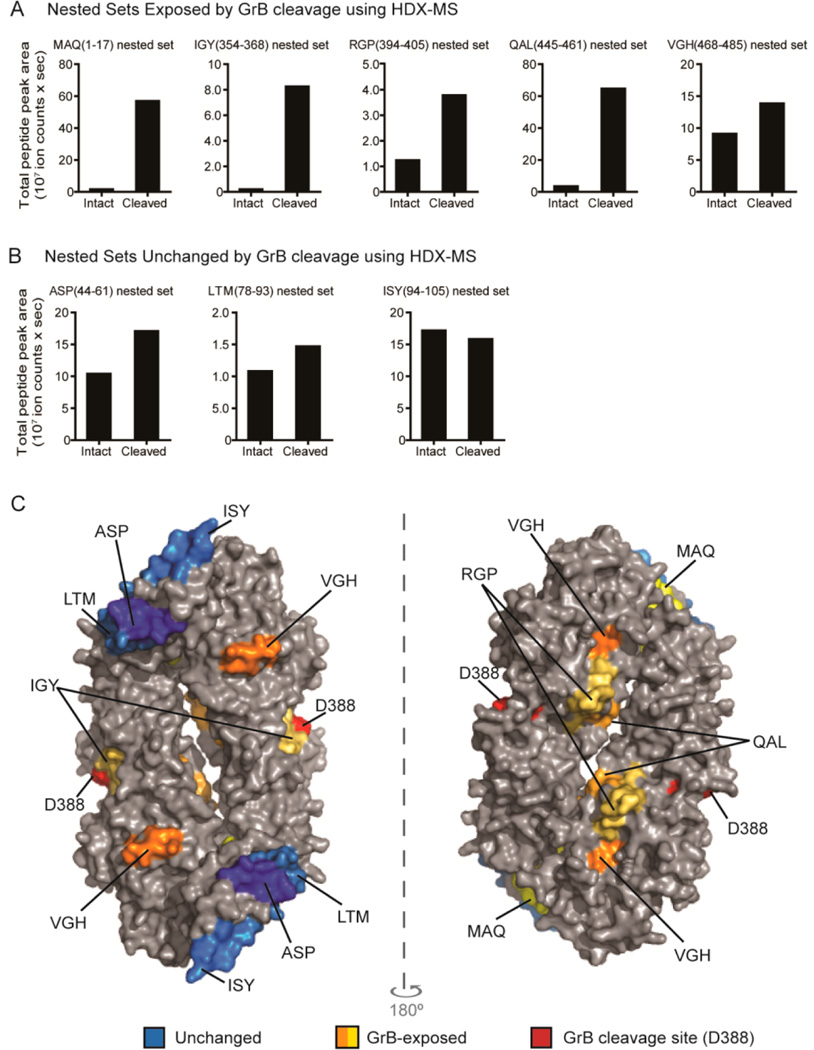
In general, MHC class II eluted peptides tend to exist as nested sets in which peptides share a common core sequence but have variable lengths at the N-and C-termini due to the open nature of the MHC class II peptide binding groove.37 When intact or GrB-cleaved PAD4 was processed by the cell-free antigen processing system, nested sets of peptides comprising eight core PAD4 epitopes were identified: MAQ(1–17), ASP(44–61), LTM(78–93), ISY(94–105), IGY(354–368), RGP(394–405), QAL(445–461), and VGH(468–485) (Table 1). For determining if GrB cleavage of PAD4 affected either the number of unique peptide sequences or the amount of specific peptide sequences presented by DR1, peptides belonging to the nested sets were identified and quantified. The relative quantity of each detected peptide sequence was determined by normalizing the area under the peptide peak to a nested set of DR1-derived peptides as an internal control for the amount of DR1 added to each sample (Table S1).
Table 1.
Nested PAD4 Peptide Sets Identified by the Cell-Free MHC Class II Antigen Processing Systema
| intact PAD4 | GrB-cleaved PAD4 | cleaved/intact PAD4 | ||||
|---|---|---|---|---|---|---|
| peptide (residues) | PAD4 peptide sequence common core epitope |
total peptide peak area (ions × sec) |
# unique peptides |
total peptide peak area (ions × sec) |
# unique peptides |
fold-change in peptide peak area |
| MAQ(1−17) | MAQGTLIRVTPEQPTHA | 1.64 × 107 | 5 | 56.94 × 107 | 20 | 34.7 |
| ASP(44−61) | ASPGVVVDIAHSPPAKKK | 10.43 × 107 | 10 | 17.07 × 107 | 11 | 1.64 |
| LTM(78−93) | LTMKAASGSTGDQKVQ | 1.08 × 107 | 2 | 1.47 × 107 | 3 | 1.36 |
| ISY(94−105) | ISYYGPKTPPVK | 17.18 × 107 | 3 | 15.84 × 107 | 3 | 0.92 |
| IGY(354−368) | IGYIQAPHKTLPVVF | 0.21 × 107 | 1 | 8.27 × 107 | 4 | 39.4 |
| RGP(394−405) | RGPQTGGISGLD | 1.24 × 107 | 2 | 3.78 × 107 | 3 | 3.05 |
| QAL(445−461) | QALQDFLSAQQVQAPVK | 3.59 × 107 | 5 | 64.74 × 107 | 10 | 18.0 |
| VGH(468−488) | VGHVDEFLSFVPAPDRKG | 9.11 × 107 | 11 | 13.88 × 107 | 12 | 1.52 |
Rows 1 and 5−7 are PAD4 peptides who presentation was enhanced >3-fold by GrB cleavage.
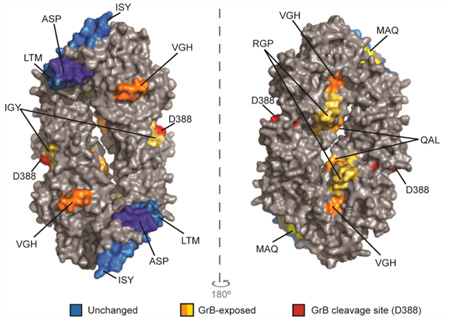
Analysis of the PAD4 peptides revealed that GrB-cleavage of PAD4 led to a 2- to 4-fold increase in the number of unique peptide sequences belonging to the MAQ, IGY, and QAL sets (Table 1 and Tables S2, S6, and S8). Proteolysis by GrB also increased the relative amount of specific peptide sequences in the MAQ, IGY, RGP, and QAL nested sets by 3- to 39-fold (Figure 4A, Table 1, and Tables S2 and S6–8). These are all derived from regions shown to undergo changes in structure by HDX following cleavage by GrB (Figure 2B). In contrast, presentation of peptides in the ASP, LTM, and ISY nested sets, which did not undergo structural changes after cleavage by GrB, was less affected (Figure 4B and Table S3–5). Presentation of peptides in the VGH nested set was also minimally affected despite this region becoming exposed following cleavage with GrB (Figure 4A and Table S9). Importantly, PAD4 epitopes, which demonstrated the most dramatic increases in antigen presentation following cleavage with GrB, colocalized to regions that were shown by HDX to be exposed following cleavage. These epitopes were located at the newly created N- and C-termini generated by GrB cleavage as well as on the inner surface of the PAD4 dimer (Figure 4C). The peptides whose presentation was not affected by GrB were derived from regions on the outside surface of the PAD4 dimer, predominantly in the N-terminal domain. These data suggest a direct role for GrB-induced structural changes in increasing presentation of cryptic PAD4 peptides.
Figure 4.
GrB cleavage of PAD4 enhanced the presentation of GrB-exposed epitopes. The normalized amount of PAD4 peptides in each nested set derived from intact PAD4 or GrB-cleaved PAD4 using the cell-free system is shown as “total peak area”. Nested sets were grouped according to whether they were derived from regions of PAD4 that were exposed by GrB cleavage (A) or remained structurally unchanged following GrB cleavage (B) using HDX. (C) The peptides identified by the cell-free system are indicated on a surface rendering of the PAD4 dimer (PBD ID: 1WDA)34 using PyMOL software. Peptides derived from regions that did not change following GrB cleavage by HDX are indicated as “unchanged” (blue area). Peptides that were derived from regions that changed with GrB cleavage are indicated as “GrB-exposed” (orange and yellow areas). The GrB cleavage site is shown (red area).
RA Patients Have T Cells Recognizing GrB-Exposed PAD4 Peptides
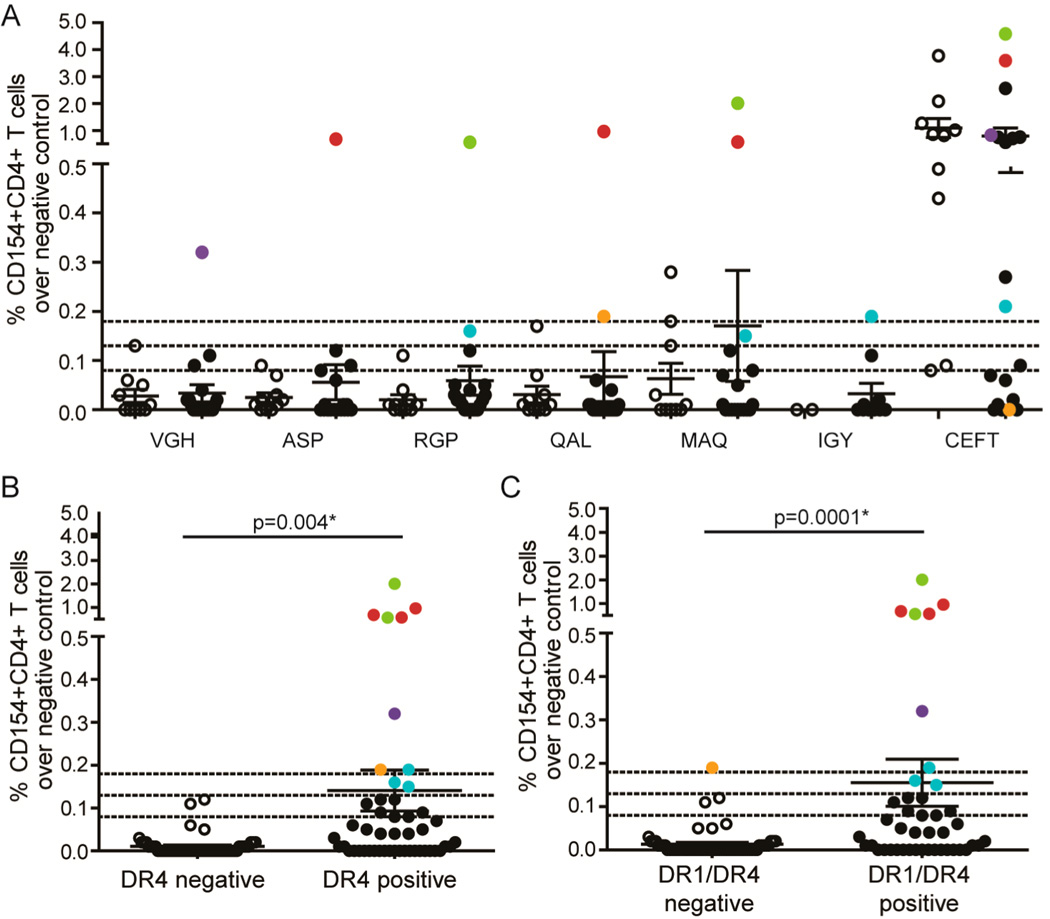
GrB-induced alterations in PAD4 structure and enhanced presentation of PAD4 peptides may be important for breaking immune tolerance to PAD4 in patients with RA. Although these initiating events likely take place early in disease prior to the onset of RA symptoms, it is only possible to study these patients once clinical disease is apparent, often years after these events have occurred.3 In this regard, CD4+ T cell responses to GrB-exposed PAD4 peptides may represent an immunologic record of these initiating events. For determining if this mechanism is active in patients with RA, PBMCs from RA patients (n = 19) and non-RA controls (n = 2 healthy controls, n = 2 psoriatic arthritis patients, and n = 6 scleroderma patients) were stimulated with a panel of PAD4 peptides presented by DR1 in the cell-free antigen processing system (Table 1). All patients and controls were positive for DR1 and/or the related MHC class II molecule DR4, both of which are SE alleles associated with the development of RA.
Robust upregulation of CD154, over 3 standard deviations above the mean of the non-RA controls, was observed on CD4+ T cells from 5/19 (26%) RA patients in response to PAD4 peptides and from 1/10 (10%) non-RA controls (Figure 5A). Importantly, RA patients and non-RA controls responded similarly to a pool of peptides from infectious sources (CMV, EBV, Flu, and Tetanus (CEFT)). Of the five RA patients responding to PAD4 peptides, four upregulated CD154 in response to at least one peptide whose presentation was enhanced by GrB. Despite the small sample size, analysis of available genetic, serological, and clinical characteristics revealed that the median level of C-reactive protein (CRP), an acute phase reactant that is elevated in the setting of inflammation, was more than 3-fold higher in patients with PAD4 peptide-specific T cells than that of patients who did not have robust T cell responses (median 2.1 vs 0.6 mg/L; p = 0.042) (Table 2). Interestingly, although these PAD4 peptides were identified based on binding to DR1 in the cell-free assay, all of the responding patients had DR4 alleles, and 4/5 (80%) coexpressed DR1 and DR4. The T cell responses to PAD4 peptides were significantly higher in RA patients who expressed DR4 alone (p = 0.004) or who coexpressed DR1 and DR4 (p = 0.0001) (Figure 5B and C). These data demonstrate that DR4-positive RA patients have circulating T cells that recognize PAD4 epitopes for which presentation is enhanced following proteolysis of PAD4 by GrB and represents an immunologic record of this mechanism occurring in vivo.
Figure 5.
RA patients have T cells recognizing GrB-exposed PAD4 peptides. (A) PBMCs from DR1 and/or DR4 positive patients with RA (n = 19) or non-RA disease and healthy controls (n = 6 scleroderma patients, n = 2 psoriatic arthritis patients, and n = 2 healthy controls) were stimulated with peptides identified by the cell-free antigen processing system or CEFT (a pool of peptides from infectious antigens). The % CD154+CD4+ T cells for each patient following incubation with the peptides over background stimulation to the negative control POLR3 peptide is shown. Dotted lines indicate 1 standard deviation (0.08), 2 standard deviations (0.13), and 3 standard deviations (0.18) above the mean of the non-RA controls. (B) RA patients are grouped according to the presence of DR4 (B) or DR1 and DR4 (C) alleles, and T cell responses to PAD4 peptides over the negative control are shown. Data were analyzed using a Wilcoxon rank-sum (Mann–Whitney) test and the median, IQR, and p-values are shown; p < 0.05 was considered significant.
Table 2.
Clinical Characteristics of RA Patients Grouped by PAD4 T Cell Statusa
| total (n = 19) |
group 1 T cell negative (n = 14) |
group 2 T cell positive (n = 5) |
group 1 vs group 2 p-value |
|
|---|---|---|---|---|
| gender, % female | 58 | 57 | 60 | 0.99 |
| raceb, % caucasian | 94 (n = 18) | 92 (n = 13) | 100 | 0.99 |
| age, mean years ± SD | 64 ±9 | 64 ± 10 | 65 ± 7 | 0.82 |
| RA duration, years | 18 (13−28) | 17 (13−30) | 19 (14−20) | 0.59 |
| DR1 positive, % | 74 | 71 | 80 | 0.99 |
| DR4 positive, % | 63 | 50 | 100 | 0.11 |
| DR1 and DR4 positive, % | 42 | 29 | 80 | 0.11 |
| anti-CCP, % | 100 | 100 | 100 | 0.99 |
| anti-CCPb, units | 100 (64−160) (n = 18) | 100 (64−160) (n = 13) | 100 (96−122) | 0.92 |
| anti-PAD4, % | 63 | 64 | 60 | 0.99 |
| CRPb, mg/L | 0.8 (0.4−1.6) (n = 17) | 0.6 (0.4−1.0) (n = 13) | 2.1 (1.2−3.2) (n = 4) | 0.042 |
| CDAI scoreb | 6.0 (1.0−9.5) (n = 15) | 5.6 (1.0−11.0) (n = 12) | 6.0 (1.3−6.0) (n = 3) | 0.77 |
| prednisoneb, % | 24 (n = 17) | 23 (n = 13) | 25 (n = 4) | 0.99 |
| nonbiologic DMARDsb, % | 76 (n = 17) | 77 (n = 13) | 75 (n = 4) | 0.99 |
| biologic DMARDsb, % | 76 (n = 17) | 69 (n = 13) | 100 (n = 4) | 0.52 |
Median (interquartile range) depicted unless otherwise noted.
Only patients with available data were included; n is indicated.
Anti-CCP = anticyclic citrullinated peptide antibodies; CDAI = clinical disease activity index; CRP = C-reactive protein; DMARDs = disease-modifying antirheumatic drugs including nonbiologic (methotrexate, sulfasalazine, leflunomide, and/or hydroxychloroquine) and biologic (monoclonal antibody inhibitors of TNFα, IL-6, or T cell costimulation); SD = standard deviation.
CONCLUSIONS
GrB has been implicated in the generation of autoantigens in autoimmune diseases by altering antigen processing, but there is a paucity of direct experimental evidence to support this hypothesis in human disease. In this study, the parallel and complementary proteomic approaches of HDX and a cell-free MHC class II antigen processing system converged to identify regions of the PAD4 protein that undergo structural changes and increased antigen presentation following cleavage by GrB at D388. These GrB-exposed peptides stimulated PAD4-specific CD4+ T cells present in the circulation of DR4-positive patients with RA and is the first demonstration that such cells exist in patients with RA and that GrB can expose immunogenic epitopes from an autoantigen.
The work presented here supports the emerging concept that proteolysis of autoantigens prior to normal antigen processing can enhance the presentation of cryptic epitopes.20–22 Proteolytic events mediated by proteases not present in the thymus or APCs may lead to the presentation of cryptic peptides and stimulation of autoreactive T cells at sights of inflammation. Recent studies from Mohan et al. have shown that a subset of autoreactive T cells were stimulated by a cryptic insulin peptide that was only generated at sites of tissue damage in the pancreas but not by normal processing of intact insulin protein by APCs.38,39 Similarly, the finding that RA patients have T cells that recognize GrB-exposed PAD4 peptides suggests that these T cells may be activated by PAD4 peptides uniquely generated in the RA target tissue. Importantly, GrB is upregulated by CTLs following antigen-specific stimulation but is not expressed by APCs or at high levels in the thymus where immune tolerance to PAD4 is established.40,41 However, tolerance to PAD4 may be broken in the inflamed RA joint if GrB cleaves PAD4 and exposes cryptic PAD4 epitopes capable of stimulating autoreactive T cells that have escaped deletion in the thymus. In this regard, high levels of soluble GrB and GrB-expressing cells are present in the synovial fluid of patients with RA,26,27 as are soluble PAD4 and PAD4-expressing cells.42,43 Although cleavage of PAD4 by GrB in the joints of patients with RA has not yet been demonstrated, the detection of T cells recognizing GrB-exposed PAD4 peptides suggests that this mechanism is operative in vivo. In addition to GrB cleavage, there are other factors that may influence PAD4 antigen processing and presentation including autoantibody binding,24,25 autocitrullination,44 and cleavage by other proteases known to be present in the inflamed RA joint.45 Understanding the contribution of these factors to the generation of cryptic PAD4 epitopes is a priority.
The detection of PAD4-specific T cells in patients with RA is significant given the substantial challenges to studying human CD4+ T cell responses in autoimmune diseases including the marked heterogeneity in the human population, the low precursor frequency of autoantigen-specific T cells in the circulation relative to the target tissue,46 and the immunomodulatory effects of disease-modifying antirheumatic drugs (DMARDs).47 The immunologic studies shown here were performed using circulating PBMCs from RA patients with long-standing disease that was well controlled with one or more DMARDs, including targeted biological therapies (Table 2). These therapies are designed to attenuate immune responses through inhibiting inflammatory cytokines, decreasing immune cell proliferation, preventing costimulation of T cells, and decreasing inflammation.47 Furthermore, it is known that T cell responses evolve over the course of disease, and epitope spreading occurs in which additional epitopes from a given antigen are recognized.48 Thus, the T cells involved in breaking tolerance to a particular autoantigen like PAD4 may not be required later to propagate established disease. These factors may have contributed to the heterogeneity in responses observed between individual patients and lack of correlation between anti-PAD4 antibodies and PAD4-specific T cells. Despite these limitations, the detection of T cells recognizing GrB-exposed PAD4 peptides suggests that enhanced presentation of PAD4 peptides by GrB has occurred in a subset of patients with RA. Future studies utilizing PBMCs from DR4 positive patients with early RA, prior to the initiation of immune modulatory therapies, are needed to study events closer to disease initiation and determine the relationship of PAD4-specific T cells to serological and clinical findings.
The finding that a subset of DR4-positive RA patients have circulating CD4+ T cells specific for GrB-exposed PAD4 peptides suggests a mechanistic role for GrB in generating cryptic PAD4 epitopes. This study has implications for numerous other autoimmune diseases in which commonly targeted autoantigens are substrates for GrB.11 It has been hypothesized that regulation of pathogenic T cells responsible for breaking tolerance early in disease may result in collapse of the autoimmune network and amelioration of disease.48 Indeed, in murine models of autoimmune diseases, nanoparticles targeting antigen-specific T cells induced an anti-inflammatory phenotype in disease-associated T, B, and dendritic cells and attenuated disease.49 Thus, defining mechanisms that result in the early steps of pathogenic T cell activation may not only have important implications for disease monitoring but could also assist in the development of antigen-specific immunotherapy.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the clinical research coordinators Michelle K. Jones, Brandy Miles, and Margaret Sampedro past and present physicians at the Johns Hopkins Arthritis Center, and Ami A. Shah, M.D., M.H.S. from the Johns Hopkins Scleroderma Center for recruiting patients for the study; Raffaello Cimbro from the Bayview Immunomics Core (BIC) flow cytometry core for his technical contribution to this work; and Tory P. Johnson, Ph.D. for critical review of this manuscript. This work was supported by the Jerome L. Greene Foundation (E.D.), the Sibley Memorial Hospital Mackley Fund (E.D., A.R.), the Johns Hopkins Arthritis Center Discovery Fund (C.O.B.), the Camille Julia Morgan Arthritis Research and Education Fund (C.O.B.), the Johns Hopkins Institute for Clinical and Translational Research UL1 TR001079 (R.N.C.), and the National Institutes of Health P30-AR053503 (A.R., C.O.B.), MH084512 (P.R.G.), GM084041 (P.R.G.), R01AI063764 (S.S.-N.), and R21AI101987 (S.S.-N.).
ABBREVIATIONS
- APC
antigen-presenting cell
- CCP
cyclic citrullinated peptide antibodies
- CDAI
clinical disease activity index
- CID
collision-induced dissociation
- CRP
c-reactive protein
- CTL
cytotoxic T lymphocyte
- DMARDs
disease-modifying antirheumatic drugs
- DR1
HLA-DR1
- DR4
HLA-DR4
- GrB
granzyme B
- HDX
hydrogen–deuterium exchange
- IQR
interquartile range
- LC–MS
liquid chromatography/mass spectrometry
- LC–MS/MS
liquid chromatography/tandem mass spectrometry
- MHC
major histocompatibility
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- PAD4
peptidyl arginine deiminase IV
- PBMCs
peripheral blood mononuclear cells
- PDB
protein data bank
- RA
rheumatoid arthritis
- SE
shared epitope
- SD
standard deviation
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Detailed mass spectrometry methods for MS/MS peptide mapping and the cell-free antigen processing system; Figure S1, complete PAD4 peptide coverage map for HDX; Figure S2, HDX profile of intact PAD4; Figure S3, additional HDX graphs for individual PAD4 peptides; Table S1–9, relative quantification of nested set peptide peak areas from intact and GrB-cleaved PAD4 (PDF)
Author Contributions
E.D. purified PAD4 protein, performed proteolysis experiments, processed and performed studies with human samples, generated the protein structure images, analyzed clinical and HLA-typing data, and was involved in the overall study design and data analysis/interpretation. A.F. was involved in developing and performing the flow cytometry experiments to measure T cell activation. A.K. purified DR1 protein, performed the cell-free system, and analyzed the resulting mass spectrometry data. S.S.-N. guided cell-free MHC class II antigen processing studies and data analysis/interpretation. T.B. performed mass spectrometry and quantification of peptides indentified by the cell-free system. R.N.C. guided mass-spectrometry analysis and troubleshooting for the cell-free assay. X.Z. designed the HDX study, performed the HDX experiments, and analyzed the HDX data. M.J.C. contributed to study design and HDX platform development. P.R.G. guided HDX study design and data analysis/interpretation. C.O.B. was responsible for patient data collection, specimen collection, and clinical data analysis of patients currently followed at the Johns Hopkins Arthritis Center. J.T.G. was responsible for patient data collection, specimen collection, and clinical data analysis of patients that were enrolled in the ESCAPE RA cohort. A.R. guided the overall study design and data analysis/interpretation. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): E.D., J.T.G., and A.R. are authors on issued patent no. 8,975,033 entitled "Human autoantibodies specific for pad3 which are cross-reactive with pad4 and their use in the diagnosis and treatment of rheumatoid arthritis and related diseases, and E.D. previously served on the scientific advisory board for Padlock Therapeutics, Inc.
REFERENCES
- 1.Engelhorn ME, Guevara-Patino JA, Noffz G, Hooper AT, Lou O, Gold JS, Kappel BJ, Houghton AN. Autoimmunity and tumor immunity induced by immune responses to mutations in self. Nat. Med. 2006;12:198–206. doi: 10.1038/nm1363. [DOI] [PubMed] [Google Scholar]
- 2.Joseph CG, Darrah E, Shah AA, Skora AD, Casciola-Rosen LA, Wigley FM, Boin F, Fava A, Thoburn C, Kinde I, Jiao Y, Papadopoulos N, Kinzler KW, Vogelstein B, Rosen A. Association of the autoimmune disease scleroderma with an immunologic response to cancer. Science. 2014;343:152–157. doi: 10.1126/science.1246886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arend WP, Firestein GS. Pre-rheumatoid arthritis: predisposition and transition to clinical synovitis. Nat. Rev. Rheumatol. 2012;8:573–586. doi: 10.1038/nrrheum.2012.134. [DOI] [PubMed] [Google Scholar]
- 4.Scally SW, Petersen J, Law SC, Dudek NL, Nel HJ, Loh KL, Wijeyewickrema LC, Eckle SB, van Heemst J, Pike RN, McCluskey J, Toes RE, La Gruta NL, Purcell AW, Reid HH, Thomas R, Rossjohn J. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J. Exp. Med. 2013;210:2569–2582. doi: 10.1084/jem.20131241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casciola-Rosen L, Miagkov A, Nagaraju K, Askin F, Jacobson L, Rosen A, Drachman DB. Granzyme B: evidence for a role in the origin of myasthenia gravis. J. Neuroimmunol. 2008;201–202:33–40. doi: 10.1016/j.jneuroim.2008.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter CR, Sayers TJ, Wiltrout RH, Turcovski-Corrales SM, Taub DD. Generation of antigenic peptides by lymphocyte granule serine proteases (granzymes) Cell. Immunol. 1996;172:235–245. doi: 10.1006/cimm.1996.0238. [DOI] [PubMed] [Google Scholar]
- 7.Dudek NL, Maier S, Chen ZJ, Mudd PA, Mannering SI, Jackson DC, Zeng W, Keech CL, Hamlin K, Pan ZJ, Davis-Schwarz K, Workman-Azbill J, Bachmann M, McCluskey J, Farris AD. T cell epitopes of the La/SSB autoantigen in humanized transgenic mice expressing the HLA class II haplotype DRB1*0301/DQB1*0201. Arthritis Rheum. 2007;56:3387–3398. doi: 10.1002/art.22870. [DOI] [PubMed] [Google Scholar]
- 8.Casciola-Rosen L, Andrade F, Ulanet D, Wong WB, Rosen A. Cleavage by granzyme B is strongly predictive of autoantigen status: implications for initiation of autoimmunity. J. Exp. Med. 1999;190:815–826. doi: 10.1084/jem.190.6.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casciola-Rosen L, Miagkov A, Nagaraju K, Askin F, Jacobson L, Rosen A, Drachman DB. Granzyme B: evidence for a role in the origin of myasthenia gravis. J. Neuroimmunol. 2008;201–202:33–40. doi: 10.1016/j.jneuroim.2008.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhury D, Lieberman J. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu. Rev. Immunol. 2008;26:389–420. doi: 10.1146/annurev.immunol.26.021607.090404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darrah E, Rosen A. Granzyme B cleavage of autoantigens in autoimmunity. Cell Death Differ. 2010;17:624–632. doi: 10.1038/cdd.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernando MM, Stevens CR, Walsh EC, De Jager PL, Goyette P, Plenge RM, Vyse TJ, Rioux JD. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet. 2008;4:e1000024. doi: 10.1371/journal.pgen.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skapenko A, Leipe J, Lipsky PE, Schulze-Koops H. The role of the T cell in autoimmune inflammation. Arthritis Res. Ther. 2005;7(Suppl 2):S4–S14. doi: 10.1186/ar1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sercarz EE, Maverakis E. Mhc-guided processing: binding of large antigen fragments. Nat. Rev. Immunol. 2003;3:621–629. doi: 10.1038/nri1149. [DOI] [PubMed] [Google Scholar]
- 15.Sadegh-Nasseri S, Kim A. Exogenous antigens bind MHC class II first, and are processed by cathepsins later. Mol. Immunol. 2015;68:81–84. doi: 10.1016/j.molimm.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antoniou AN, Blackwood SL, Mazzeo D, Watts C. Control of antigen presentation by a single protease cleavage site. Immunity. 2000;12:391–398. doi: 10.1016/s1074-7613(00)80191-0. [DOI] [PubMed] [Google Scholar]
- 17.Manoury B, Mazzeo D, Fugger L, Viner N, Ponsford M, Streeter H, Mazza G, Wraith DC, Watts C. Destructive processing by asparagine endopeptidase limits presentation of a dominant T cell epitope in MBP. Nat. Immunol. 2002;3:169–174. doi: 10.1038/ni754. [DOI] [PubMed] [Google Scholar]
- 18.Kim A, Hartman IZ, Poore B, Boronina T, Cole RN, Song N, Ciudad MT, Caspi RR, Jaraquemada D, Sadegh-Nasseri S. Divergent paths for the selection of immunodominant epitopes from distinct antigenic sources. Nat. Commun. 2014;5:5369. doi: 10.1038/ncomms6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sercarz EE, Lehmann PV, Ametani A, Benichou G, Miller A, Moudgil K. Dominance and crypticity of T cell antigenic determinants. Annu. Rev. Immunol. 1993;11:729–766. doi: 10.1146/annurev.iy.11.040193.003501. [DOI] [PubMed] [Google Scholar]
- 20.Lanzavecchia A. How can cryptic epitopes trigger autoimmunity? J. Exp. Med. 1995;181:1945–1948. doi: 10.1084/jem.181.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moudgil KD, Sercarz EE. Understanding crypticity is the key to revealing the pathogenesis of autoimmunity. Trends Immunol. 2005;26:355–359. doi: 10.1016/j.it.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 22.Sadegh-Nasseri S, Kim A. MHC Class II Auto-Antigen Presentation is Unconventional. Front. Immunol. 2015;6:372. doi: 10.3389/fimmu.2015.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. BioEssays. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 24.Harris ML, Darrah E, Lam GK, Bartlett SJ, Giles JT, Grant AV, Gao P, Scott WW, Jr, El-Gabalawy H, Casciola-Rosen L, Barnes KC, Bathon JM, Rosen A. Association of autoimmunity to peptidyl arginine deiminase type 4 with genotype and disease severity in rheumatoid arthritis. Arthritis Rheum. 2008;58:1958–1967. doi: 10.1002/art.23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darrah E, Giles JT, Ols ML, Bull HG, Andrade F, Rosen A. Erosive rheumatoid arthritis is associated with antibodies that activate PAD4 by increasing calcium sensitivity. Sci. Transl. Med. 2013;5:186ra65. doi: 10.1126/scitranslmed.3005370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tak PP, Kummer JA, Hack CE, Daha MR, Smeets TJ, Erkelens GW, Meinders AE, Kluin PM, Breedveld FC. Granzyme-positive cytotoxic cells are specifically increased in early rheumatoid synovial tissue. Arthritis Rheum. 1994;37:1735–1743. doi: 10.1002/art.1780371205. [DOI] [PubMed] [Google Scholar]
- 27.Tak PP, Spaeny-Dekking L, Kraan MC, Breedveld FC, Froelich CJ, Hack CE. The levels of soluble granzyme A and B are elevated in plasma and synovial fluid of patients with rheumatoid arthritis (RA) Clin. Exp. Immunol. 1999;116:366–370. doi: 10.1046/j.1365-2249.1999.00881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraan MC, Haringman JJ, Weedon H, Barg EC, Smith MD, Ahern MJ, Smeets TJ, Breedveld FC, Tak PP. T cells, fibroblast-like synoviocytes, and granzyme B+ cytotoxic cells are associated with joint damage in patients with recent onset rheumatoid arthritis. Ann. Rheum. Dis. 2004;63:483–488. doi: 10.1136/ard.2003.009225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldbach-Mansky R, Suson S, Wesley R, Hack CE, El-Gabalawy HS, Tak PP. Raised granzyme B levels are associated with erosions in patients with early rheumatoid factor positive rheumatoid arthritis. Ann. Rheum. Dis. 2005;64:715–721. doi: 10.1136/ard.2003.007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carson MW, Zhang J, Chalmers MJ, Bocchinfuso WP, Holifield KD, Masquelin T, Stites RE, Stayrook KR, Griffin PR, Dodge JA. HDX reveals unique fragment ligands for the vitamin D receptor. Bioorg. Med. Chem. Lett. 2014;24:3459–3463. doi: 10.1016/j.bmcl.2014.05.070. [DOI] [PubMed] [Google Scholar]
- 31.Hartman IZ, Kim A, Cotter RJ, Walter K, Dalai SK, Boronina T, Griffith W, Lanar DE, Schwenk R, Krzych U, Cole RN, Sadegh-Nasseri S. A reductionist cell-free major histocompatibility complex class II antigen processing system identifies immunodominant epitopes. Nat. Med. 2010;16:1333–1340. doi: 10.1038/nm.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Chien EY, Chalmers MJ, Pascal BD, Gatchalian J, Stevens RC, Griffin PR. Dynamics of the beta(2)-Adrenergic G-Protein Coupled Receptor Revealed by Hydrogen-Deuterium Exchange. Anal. Chem. 2010;82:1100–1108. doi: 10.1021/ac902484p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pascal BD, Chalmers MJ, Busby SA, Griffin PR. HD desktop: an integrated platform for the analysis and visualization of H/D exchange data. J. Am. Soc. Mass Spectrom. 2009;20:601–610. doi: 10.1016/j.jasms.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. Structural basis for Ca(2+)-induced activation of human PAD4. Nat. Struct. Mol. Biol. 2004;11:777–783. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 35.Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J. Am. Soc. Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landry SJ. Local protein instability predictive of helper T-cell epitopes. Immunol. Today. 1997;18:527–532. doi: 10.1016/s0167-5699(97)01152-3. [DOI] [PubMed] [Google Scholar]
- 37.Chicz RM, Urban RG, Lane WS, Gorga JC, Stern LJ, Vignali DA, Strominger JL. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature. 1992;358:764–768. doi: 10.1038/358764a0. [DOI] [PubMed] [Google Scholar]
- 38.Mohan JF, Levisetti MG, Calderon B, Herzog JW, Petzold SJ, Unanue ER. Unique autoreactive T cells recognize insulin peptides generated within the islets of Langerhans in autoimmune diabetes. Nat. Immunol. 2010;11:350–354. doi: 10.1038/ni.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohan JF, Calderon B, Anderson MS, Unanue ER. Pathogenic CD4(+) T cells recognizing an unstable peptide of insulin are directly recruited into islets bypassing local lymph nodes. J. Exp. Med. 2013;210:2403–2414. doi: 10.1084/jem.20130582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gershenfeld HK, Hershberger RJ, Shows TB, Weissman IL. Cloning and chromosomal assignment of a human cDNA encoding a T cell- and natural killer cell-specific trypsin-like serine protease. Proc. Natl. Acad. Sci. U. S. A. 1988;85:1184–1188. doi: 10.1073/pnas.85.4.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meng Y, Harlin H, O’Keefe JP, Gajewski TF. Induction of cytotoxic granules in human memory CD8+ T cell subsets requires cell cycle progression. J. Immunol. 2006;177:1981–1987. doi: 10.4049/jimmunol.177.3.1981. [DOI] [PubMed] [Google Scholar]
- 42.Foulquier C, Sebbag M, Clavel C, Chapuy-Regaud S, Al Badine R, Mechin MC, Vincent C, Nachat R, Yamada M, Takahara H, Simon M, Guerrin M, Serre G. Peptidyl arginine deiminase type 2 (PAD-2) and PAD-4 but not PAD-1, PAD-3, and PAD-6 are expressed in rheumatoid arthritis synovium in close association with tissue inflammation. Arthritis Rheum. 2007;56:3541–3553. doi: 10.1002/art.22983. [DOI] [PubMed] [Google Scholar]
- 43.Umeda N, Matsumoto I, Kawaguchi H, Kurashima Y, Kondo Y, Tsuboi H, Ogishima H, Suzuki T, Kagami Y, Sakyu T, Ishigami A, Maruyama N, Sumida T. Prevalence of soluble peptidylarginine deiminase 4 (PAD4) and anti-PAD4 antibodies in autoimmune diseases. Clin. Rheumatol. 2016;35:1181–1188. doi: 10.1007/s10067-015-3082-z. [DOI] [PubMed] [Google Scholar]
- 44.Andrade F, Darrah E, Gucek M, Cole RN, Rosen A, Zhu X. Autocitrullination of human peptidylarginine deiminase 4 regulates protein citrullination during cell activation. Arthritis Rheum. 2010;62:1630–1640. doi: 10.1002/art.27439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahrens D, Koch AE, Pope RM, Stein-Picarella M, Niedbala MJ. Expression of matrix metalloproteinase 9 (96-kd gelatinase B) in human rheumatoid arthritis. Arthritis Rheum. 1996;39:1576–1587. doi: 10.1002/art.1780390919. [DOI] [PubMed] [Google Scholar]
- 46.Bacher P, Scheffold A. New technologies for monitoring human antigen-specific T cells and regulatory T cells by flow-cytometry. Curr. Opin. Pharmacol. 2015;23:17–24. doi: 10.1016/j.coph.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 47.Bays AM, Gardner G. Pharmacologic Therapies for Rheumatologic and Autoimmune Conditions. Med. Clin. North Am. 2016;100:719–731. doi: 10.1016/j.mcna.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Sercarz EE. Driver clones and determinant spreading. J. Autoimmun. 2000;14:275–277. doi: 10.1006/jaut.2000.0380. [DOI] [PubMed] [Google Scholar]
- 49.Clemente-Casares X, Blanco J, Ambalavanan P, Yamanouchi J, Singha S, Fandos C, Tsai S, Wang J, Garabatos N, Izquierdo C, Agrawal S, Keough MB, Yong VW, James E, Moore A, Yang Y, Stratmann T, Serra P, Santamaria P. Expanding antigen-specific regulatory networks to treat autoimmunity. Nature. 2016;530:434–440. doi: 10.1038/nature16962. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.