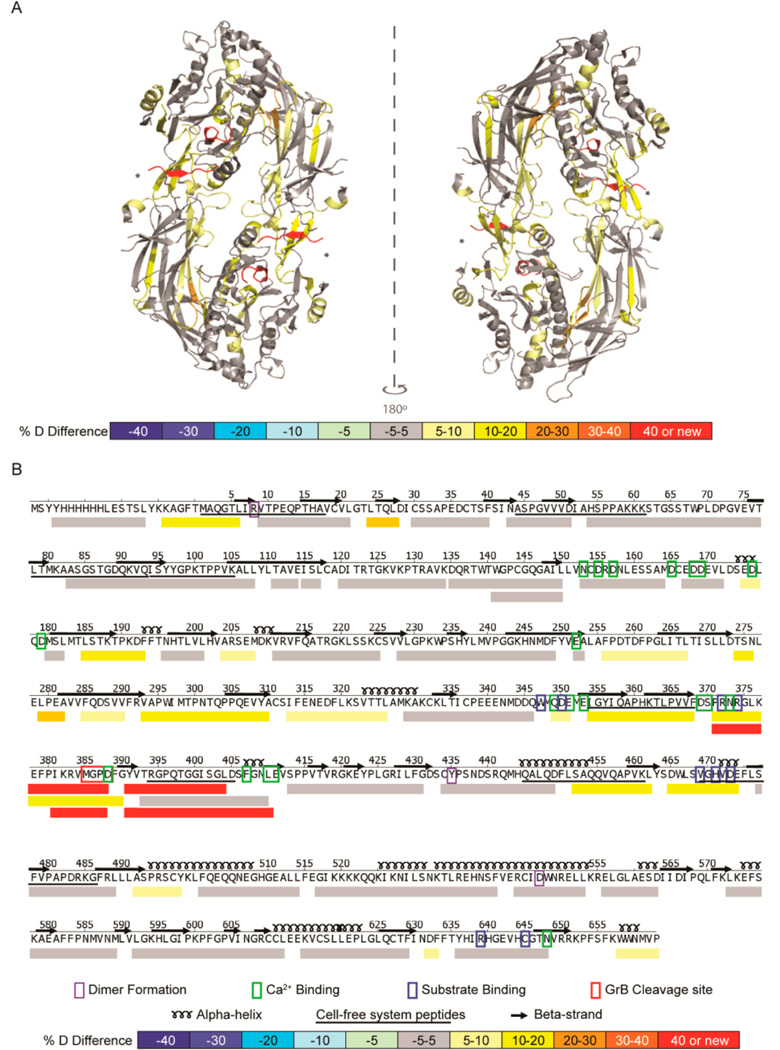
Figure 2.
GrB cleavage of PAD4 induced structural changes adjacent to and remote from the cleavage site. (A) A differential HDX profile was generated indicating regions of the PAD4 protein that were exposed following proteolysis by GrB and was superimposed onto the calcium-free PAD4 dimeric crystal structure (PDB ID: 1WD8).34 The GrB cleavage site (*) demonstrates increased structural dynamics with the formation of new N- and C-termini (shown in red). (B) The linear differential HDX profile indicates the secondary structure elements (α-helices and β-strands), residues important for dimer formation (purple box), calcium binding (green box), and substrate binding (blue box), as well as the GrB cleavage site (red box). The %D difference was calculated by subtracting the %D observed for each peptide derived from intact PAD4 from that observed from peptides derived from GrB-cleaved PAD4 and is represented by a color-coded scale. Peptides identified by the cell-free MHC class II antigen processing system are underlined. HDX–MS was performed on intact and GrB-cleaved PAD4 in quadruplicate.