Fig. 1.
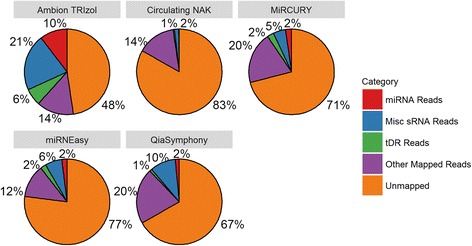
Pie chart that depicts the percentage of number of reads in different alignment categories: miRNA (include isomiR), tDR, osRNA, other mapped reads, unmapped. For any sRNA sequencing project on tissue, the unmapped rate will be at least 50%. The unmapped rate will be even higher for extracellular sRNA sequencing because low sRNA content in serum
