Fig. 1.
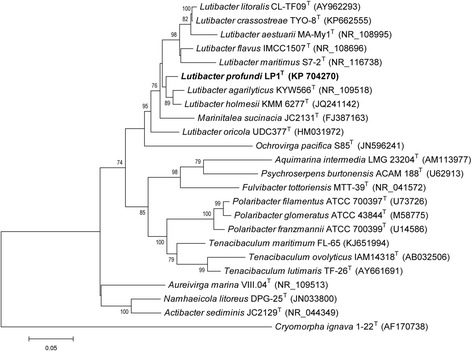
Phylogenetic tree displays the position of Lutibacter profundi LP1T (shown in bold) relative to the other type strains of Lutibacter based on 16S rRNA. The phylogenetic tree was generated after trimming the alignment using MUSCLE [82, 83] to 1323 aligned positions, using maximum likelihood method with general time reversible model as preferred model incorporated in MEGA v. 6.06 [84]. At the branch points bootstrap values above 70, expressed as percentage of 1000 replicates, are shown. Bar: 0.05 substitutions per nucleotide position. Cryomorpha ignava 1-22T (AF170738) was used as outgroup
